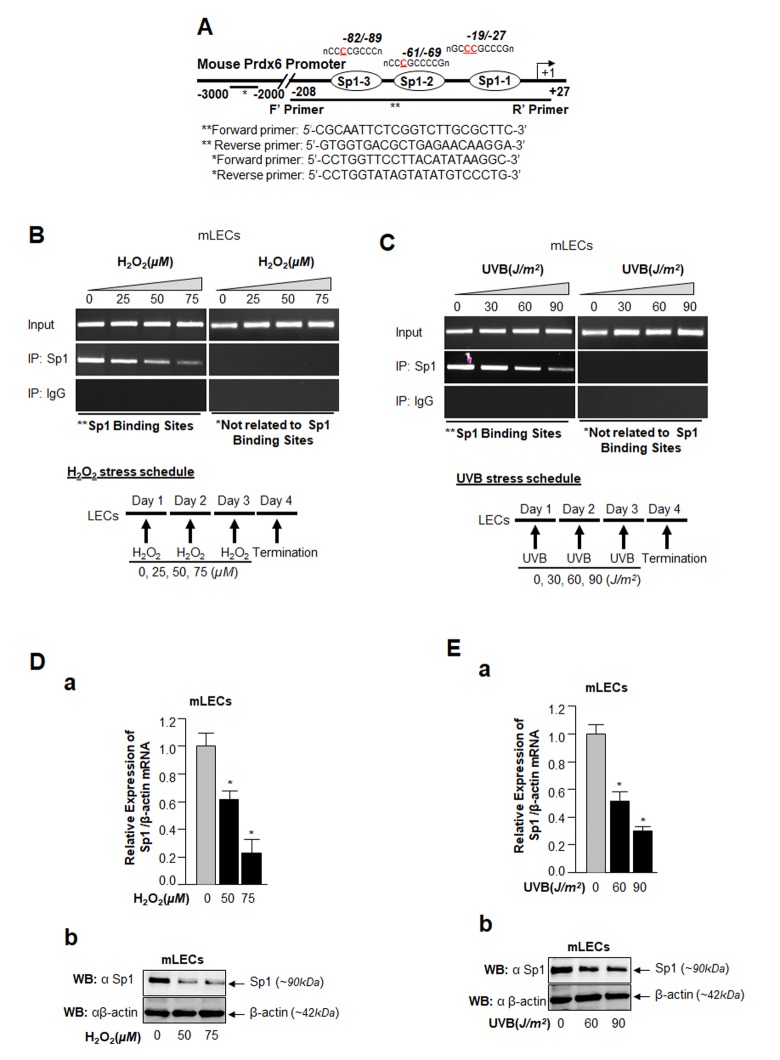
Figure 2.
ChIP analysis of genomic DNA from LECs facing oxidative stress disclosed a significant loss in Sp1-DNA binding to Prdx6 gene promoter. (A) Schematic illustration of 5’-proximal promoter region of Prdx6 containing Sp1 binding sites showing primer location and sequences used in ChIP assay. (B and C) ChIP assay showing Sp1 binding to Prdx6 promoter in vivo. Chromatin samples were prepared from Prdx6+/+ LECs (mLECs) exposed to different doses of H2O2 (0, 25, 50 and 75µM) and/or UVB (0, 30, 60 and 90J/m2) as indicated. 72h later samples were subjected to ChIP assay with ChIP grade antibodies, anti-Sp1 or IgG control. The DNA fragments were amplified by using primers designed to amplify −208 to +27 region of the Prdx6 promoter bearing Sp1 sites (**) and contiguous sequence (−2229 to −2356) to which Sp1 does not bind (*) as indicated. PCR products were resolved onto agarose gel and visualized with ethidium bromide staining. Photographs are representative of three experiments. (D and E) Expression assays showing H2O2- and UVB- induced declined expression of Sp1 in mLECs. mLECs cells were treated with different concentrations of H2O2 (D) and/or UVB (E) multiple time for 3 days as indicated. Total RNA and protein were isolated and subjected to real-time PCR and Western analysis with Sp1 specific probes, respectively. Data revealed a concentration –dependent reduced expression of Sp1 mRNA (Da and Ea; Gray vs black bars; *p<0.001) and protein (Db and Eb).