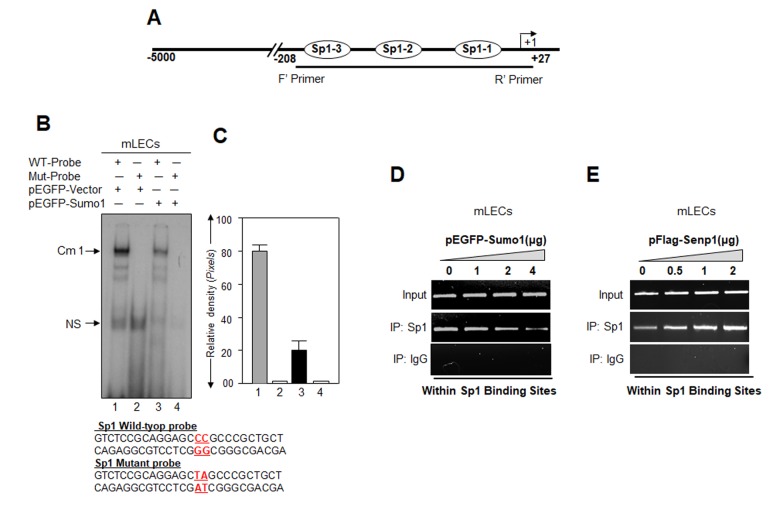
Figure 6.
Cells overexpressing Sumo1 showed reduced Sp1 binding to its responsive elements in Prdx6 promoter. (A) Schematic illustration of Prdx6 gene promoter. (B) Gel-shift mobility assay showing that Sumo1 reduced the Sp1 DNA–binding activity of Prdx6 gene promoter. Gel-shift mobility assay was carried out using nuclear extracts isolated from mLECs transfected with pEGFP-Vector (Lanes 1 and 2) or pEGFP-Sumo1 (Lanes 3 and 4) incubated with 32p-labeled wild type probe (Lanes 1 and 3) or its mutant (Lanes 2 and 4). A diminished Cm1 band was observed in cells overexpressing Sumo1 (Lane 3) in comparison to vector control (Lane 1). No binding occurred in mutant probes (Lanes 2 and 4). (C) Histogram represents densitometry analysis of DNA-protein complex formed in gel-shift assay. Lane 1 vs lane 3, *p < 0.001. (D) ChIP assay showing Sumo1 overexpression significantly suppressed Sp1-DNA binding in Prdx6 gene promoter in dose-dependent manner. mLECs were transiently transfected with different concentrations of pEGFP-Sumo1 (0, 1, 2 and 4 μg). 72h later ChIP assay was carried out with anti-Sp1 and control IgG antibodies. Pulled DNA fragments were subjected to PCR analysis for Sp1 binding cis-elements of Prdx6 promoter. The product was analyzed through agarose-gel. Data represent three experiments. (E) Senp1 overexpression dramatically enhanced Sp1-DNA binding in concentration-dependent -manner. mLECs were transfected with increasing concentrations of pFlag-Senp1 (0, 0.5, 1 and 2μg) for 72h. ChIP analysis was carried out using chromatin samples prepared from transfected LECs with a ChIP grade antibody, anti-Sp1 and control IgG. The DNA fragments were used as templates for PCR by using primers designed to amplify −208 to +27 region of the Prdx6 promoter bearing Sp1 binding sites as shown. PCR product was analyzed through agarose gel as shown.