Abstract
The latent structure of schizotypy and psychosis-spectrum symptoms remains poorly understood. Furthermore, molecular genetic substrates are poorly defined, largely due to the substantial resources required to collect rich phenotypic data across diverse populations. Sample sizes of phenotypic studies are often insufficient for advanced structural equation modeling approaches. In the last 50 years, efforts in both psychiatry and psychological science have moved toward (1) a dimensional model of psychopathology (eg, the current Hierarchical Taxonomy of Psychopathology [HiTOP] initiative), (2) an integration of methods and measures across traits and units of analysis (eg, the RDoC initiative), and (3) powerful, impactful study designs maximizing sample size to detect subtle genomic variation relating to complex traits (the Psychiatric Genomics Consortium [PGC]). These movements are important to the future study of the psychosis spectrum, and to resolving heterogeneity with respect to instrument and population. The International Consortium of Schizotypy Research is composed of over 40 laboratories in 12 countries, and to date, members have compiled a body of schizotypy- and psychosis-related phenotype data from more than 30000 individuals. It has become apparent that compiling data into a protected, relational database and crowdsourcing analytic and data science expertise will result in significant enhancement of current research on the structure and biological substrates of the psychosis spectrum. The authors present a data-sharing infrastructure similar to that of the PGC, and a resource-sharing infrastructure similar to that of HiTOP. This report details the rationale and benefits of the phenotypic data collective and presents an open invitation for participation.
Keywords: data sharing, schizotypy, schizotypal, psychosis, schizophrenia, phenotype, genetic, ICSR, HiTOP
Recent progress in psychiatric and psychological science underscores the need for consolidation and meta-analysis of phenotypic and molecular data, to model the latent structure of the psychosis spectrum. Support for this undertaking stems from the inadequacy of categorical diagnoses alone to reflect the apparent spectrum of psychotic disorders, quickly developing dimensional conceptualizations of psychopathology,1 the high polygenicity of psychosis symptom dimensions2 (also T. B. Bigdeli et al, unpublished data), and the selective role of rare variants in conferring risk for psychosis.3–5
Three initiatives, proceeding largely independently, have brought the field toward a critical juncture in which consolidation efforts are likely to significantly improve our understanding of severe psychopathology: the Psychiatric Genomics Consortium (PGC) has enhanced our genetic understanding of the psychosis spectrum,6–29 the Hierarchical Taxonomy of Psychopathology consortium (HiTOP) has endeavored to map the latent structure of psychosis,1,30 and the National Institute of Mental Health (NIMH) Research Domain Criteria (RDoC) initiative has endeavored to develop crosswalks between multiple units of analyses (eg, behavioral paradigms relevant to schizotypy measures).31–38 Consolidation efforts are also consistent with the translational aims of the Roadmap for Mental Health Research in Europe (ROAMER) project.39
As van Os et al40 have recently asserted, the psychosis spectrum requires careful reconstruction. The International Consortium for Schizotypy Research (ICSR) convened in March of 2017 to discuss current research and ways to improve understanding of dimensionality and discontinuity in schizotypy and risk for psychosis. The steering committee moved to collectively amass data and secured collaborations with the PGC and HiTOP to ensure informed data consolidation, reflecting strategies implemented by the PGC.41 This report details further the rationale for data sharing, the advantages it provides to collaborators, and the process by which we hope to achieve PGC-, HiTOP-, and RDoC-related aims. Broadly, the current goal of the ICSR is to create a data resource that will continue to grow and lead to discoveries which inform biology and nosology, improve assessment, and identify treatment targets.
Rationale for ICSR Data Sharing
There are several important reasons to amass phenotypic data on the psychosis spectrum. There is some consensus that the current concept of “schizophrenia,” described by diagnostic guidelines and later reified, confines research to a constantly changing “construct that does not exist.”42 Research on schizotypy, ie, the latent diathesis for psychosis and psychosis-spectrum disorders,43 and schizotypal signs and symptoms has addressed some of the problems of “reification” by characterizing cognitive and emotional facets of these symptoms across populations,44–55 and by comparing categorical high-risk states with symptoms in nonclinical, healthy populations. But because categorical conceptualizations of high-risk states and psychometrically-identified schizotypy can be similarly “reified,” symptom dimensions should be empirically evidenced and mapped more comprehensively across a broad network of phenotypes, with careful consideration of the differences between phenomena and symptoms as well as assessments. Due to the heterogeneous nature of assessments, limited sampling and insufficient statistical power to conduct appropriate structural equation modeling, this issue must be addressed with mass collaboration.
Individuals Identified by Current Psychometric Approaches Appear to Represent a Small Fraction of a Heterogeneous Spectrum Phenotype
Psychometrically identified high-risk groups, based on arbitrary cut-offs, have restricted research to the narrow view of psychotic experiences and/or extreme anhedonia (eg, “ultra-high risk”), despite evidence from genomic research that such individuals are a small sample of the psychosis spectrum. Subthreshold psychosis spectrum symptoms should be redefined and supplemented to improve prediction of actual onset of psychosis in the general population.
The structure of psychosis and related sequelae, within a hierarchical model such as HiTOP, remains relatively undefined compared with other dimensional components of personality and internalizing/externalizing disorders.1,56 Addressing this concern requires enhanced quantitative approaches to refining what we consider to be schizotypal traits, ideally involving network, longitudinal growth curve, and machine learning approaches—all of which are impossible with the currently limited availability of psychosis-spectrum phenotypic data. This also requires careful attention to the constructs involved and their conceptualization.57–60
Psychiatric Traits and Symptoms Are Genetically Complex, and Light Phenotyping Is Inherent to Large Genomic Efforts
Using very large samples collected for genomic mega-analysis, we experience the drawbacks of necessarily lighter phenotyping—dramatically abbreviated scales, or even the use of a single item—and a reduced diversity of clinical and behavioral data. It has become apparent that very small numbers of items are needed to economically test genomic relationships in large epidemiological studies, and the PGC working groups are interested in careful psychometric validation of such items. With many different datasets, measures, methods, and populations compiled for side-by-side comparison, the field is better able to identify effective items using methods such as confirmatory factor analysis (CFA) and item response theory (IRT). Moreover, items may be tailored to culture or clinical population (case, pedigree, college student). One deliverable advance stemming from this effort is developmental and testing support from involved researchers for a web application that may be used in large-scale epidemiological studies. Not only will replicable findings on the validity and utility of items be useful to the PGC, but these efforts will in turn inform phenotypic measurement.
Schizotypy may Easily Vary by Genomic Profile, and Rare Genomic Features Could Isolate Key Symptom Dimensions
Genetic subtyping of psychosis-spectrum disorders is highly desirable if it can lead to more accurate classification, early prediction, and effective pharmacological interventions.61 We observe in genomic psychosis-spectrum research that (1) traits are highly polygenic, and yet (2) specific rare variants result in psychosis despite low genome-wide polygenic risk for schizophrenia.5
Genetic subtyping of complex psychiatric traits has been slow to develop, but is moving forward in autism spectrum disorder research, where many probands in dense pedigrees inherit rare variants which disproportionally affect cognitive ability.62 It is possible that probands with rare mutations will have symptoms similar to those with more typical genetic profiles, but this is not a certainty, and the degree to which probands are atypical is facilitated by modeling the genetic and phenotypic heterogeneity of individuals with autism spectrum disorders.63 The same can be said for schizophrenia. The genome is increasingly examined as a dimensional measure (rather than inspected for genome-wide significant hits) to characterize schizophrenia risk, and this is leading to findings relevant to classification.25,64–66 These methods, although advancing, remain hindered by over-reliance on categorical diagnosis. We can facilitate informed decisions about clinically meaningful differences within the psychosis spectrum after proper symptom data consolidation efforts across multiple research groups, integrating family history, personality, and developmental data.
Dimensional Data Are Statistically Powerful, and Some Categorical Measures can Harmonize With Dimensional Measures
It is now established that schizotypal symptom expression, as currently measured, is detectable across the general population, in biological relatives, and in probands, and can be better characterized using a quantitative dimensional approach than a dichotomous distinction made with arbitrary cut points.58,67 To date, the “ultra-high risk” characterization has had moderate clinical utility, and “schizotypy” categorical distinctions have been useful insofar as measures have leveraged several phenotypes at once to gather additional evidence of dimensionality across healthy and clinical populations.68–81 Again, assumptions that the endophenotype is any less complex or heterogeneous than schizophrenia itself should be avoided,82 and can lead to premature attempts at parsing symptom factors. Reviews of the literature that integrate studies using dimensional measures have proven to be a promising start,58,83–87 but with harmonization of measures across large numbers of samples, and proper assessment of measurement invariance, we can begin to build statistically powerful models of causation and genome/environment interactions.
In psychiatry, the movement toward a dimensional framework for defining and diagnosing psychiatric illness is well established and appears to be a more reliable and comprehensive model than the Diagnostic and Statistical Manual of Mental Disorders-5 (DSM-5) and International Classification of Diseases, 10th Revision (ICD-10) categorical framework.1,88 However, psychosis-spectrum conditions prove problematic in the typical internalizing/externalizing dimensional framework, leading researchers to propose and test a distinct psychosis dimension.30,89,90
There is a need for clarity regarding how this psychosis dimension ought to be structured. One camp suggests a return to the model which preceded Kraepelin’s classification of 2 types of psychosis.91 This would create a single, unifying dimension of psychosis which encompasses both of Kraepelin’s distinctions,92 and indeed there is ample evidence to suggest that the conditions of psychosis share common genetic and environmental factors.93 However, a singular psychosis dimension may also fail to capture the complexity of a given condition. Multiple studies have found that a 5-dimensional structure, including dimensions labeled internalizing, disinhibited externalizing, antagonistic externalizing, detachment, and thought disorder, better harmonizes with existing categorical diagnoses.94–96 Yet other researchers have sought to blend the parsimony of a single dimension with the nuance of a 5-dimensional structure using a bi-factor model in which a general psychosis dimension is assessed first, and then used as a guide for assessment by the 5 specific symptom dimensions.97,98 The efforts of ICSR will further inform these findings and evaluate current proposed models.
The Clinic: Current High-Risk Classification Approaches Are Not Sufficient to Understand Risk
Field evidence has yet to justify a DSM risk syndrome category, with only 11% meeting criteria for ultra-high risk (UHR) classification developing psychosis in one study,99 and 39% in another.100 Inclusion of an attenuated psychosis syndrome into the full text of DSM-5.1 is still being debated.101 The best psychometrically guided predictors in nonclinical populations, outside of family history, are extreme scores on symptom surveys, and even they do not predict actual psychosis at high rates. Negative schizotypy studies find an impressive 24% of students with (categorical) extreme social anhedonia develop schizophrenia-spectrum psychopathology, but not typically psychosis. In general, schizotypy measures in predicting psychosis have been underwhelming (for review of this literature, see Docherty and Sponheim85). Family and molecular genetic studies have provided evidence that dimensionally measured negative symptoms may hold predictive utility55,68,80,102–105 but again, phenotyping in genomic studies thus far has been light, or samples too small, to adequately examine the genomic architecture of psychosis-related symptom dimensions.
The take-home message of this research is (1) family history predicts general psychopathology, (2) subthreshold symptoms of a psychosis-spectrum disorder predict symptoms of the disorder, (3) there is little diagnostic specificity with regard to prediction, and (4) the more prevalent the disorder, the greater role environment plays in etiology. Given these points, perpetuating the literature on clinically measured high risk without refining the phenotype is unlikely to improve research on early intervention.
One function of the ICSR can be to better operationalize phenotypes in accordance with dimensional models, and to improve understanding of the relation of schizotypy to other psychosis spectrum phenomenology, eg, of UHR and frank psychosis. Language, social behavior, emotional expression, beliefs, perceptual experience, and emotional response can all be reduced to behavioral function, and these can be normed using large international samples. Relatedly, an advantage of the effort is to explore the role of culture on illness expression and phenotype. The ICSR is well poised to accomplish this, given that it is truly international and intercultural.
The NIMH, PGC, HiTOP, and ICSR are moving toward a framework that is more compatible with dimensional biological risk, and the UHR/familial-high risk (FHR) research community is encouraged to collaborate with these efforts to improve clinical outcomes. Importantly, the addition of dimensional measures is meant to enhance our understanding of the latent structure of psychosis and psychosis risk, but not mire us in assumptions about diagnosis or dimension.
An Open Invitation
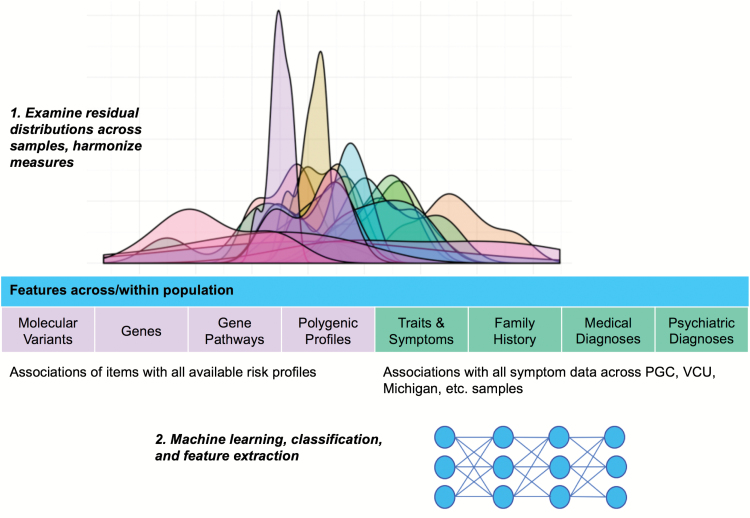
Three primary steps of this initiative are illustrated in figure 1. Data distributions are examined and large matrices of data used to develop empirically driven covariance structures. Models will implicate facets of schizotypy and psychosis-related symptoms relevant to specific intervention targets, and will be assessed relative to genomic findings. These data will include both clinical and psychometric measures. With larger sample sizes, structural equation modeling, item response theory, machine learning, and other relevant methods may be implemented to validate models of latent structure.
Fig. 1.
Overview of International Consortium of Schizotypy Research collaborative efforts.
The ICSR requires a massive, collective effort to obtain and maintain data to facilitate efficient analysis and replication. This initiative is facilitated in part by Anna Docherty’s cluster farm and by the Utah Center for Genomic Discovery (UCGD), a University of Utah initiative to integrate patient genome information into health care and develop tools for genome interpretation. Together this accounts for 2.5 PB of disc storage. There is a core team of 6 on-site data scientists and analysts, including authors Anna Docherty, John Anderson, Andrey Shabalin, and Daniel Adkins. Computing space, resources for proband and extended pedigree data collection, and an active undergraduate data collection are available to collaborative PIs. The authors of this publication themselves share broad skill sets and actively seek collaboration with other interested PIs.
“Schizotypy” is a complex, multidimensional construct, whose dimensions differ widely both in the degree and specificity with which they reflect genetic liability to schizophrenia.67 Thus to date, our consolidated data come from multiple community, risk, family, and case populations. Thirty of our collaborators share clinical and/or cognitive data from first-degree biological relatives, 19 share college student data, and 21 share community data. Most have also collected data in cases, with measures that can be harmonized and meta-analyzed. The ICSR has amassed approximately 40000 samples with clinical phenotype data including items from schizotypy and schizotypal personality assessments. We hope to double samples over the following year and plan to apply for additional external funding. These data and measures will be characterized in a first publication with all collaborators.
A portal for participation is housed on the ICSR website, at srconsortium.org. PIs or institutions may use this website to contact the data sharing facilitators. Participation includes invitation to collaborate on analyses and publication of scientific findings. Projects will be managed in the same way data analytics are handled within PGC, such that individuals and research teams submit proposals to analyze data. One example analysis by the ICSR and PGC may be focused on item refinement for future genomic research using Smartphone applications. Another study will pilot items in a genetic study of undergraduate samples.
We draw attention to the precedent of the PGC to effectively organize and collaborate on a large scale. This is done with adherence to common principles described in the inaugural publication.41 Participating members will use identical guidelines and are in active consultation with the PGC Director Patrick Sullivan to promote external review of the collaborative.
With the introduction of spectrum phenotypes in DSM-5, the field is better positioned to synthesize research to refine signs and symptoms. Impactful psychosis research in the present day reflects collective efforts to understand (1) the genetic architecture of psychosis, and (2) the latent structure of the psychosis spectrum. These efforts can be symbiotic and benefit both genomic and phenotypic research.
References
- 1. Kotov R, Krueger RF, Watson D, et al. The Hierarchical Taxonomy of Psychopathology (HiTOP): a dimensional alternative to traditional nosologies. J Abnorm Psychol. 2017;126:454–477. [DOI] [PubMed] [Google Scholar]
- 2. Docherty AR, Bigdeli TB, Edwards AC, et al. Genome-wide gene pathway analysis of psychotic illness symptom dimensions based on a new schizophrenia-specific model of the OPCRIT. Schizophr Res. 2015;164:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. [DOI] [PubMed] [Google Scholar]
- 4. Vacic V, McCarthy S, Malhotra D, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marshall CR, Howrigan DP, Merico D, et al. ; Psychosis Endophenotypes International Consortium; CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan Y, Lim ET, Sandholm N, et al. ; DIAGRAM Consortium; GENIE Consortium; GIANT Consortium; IIBDGC Consortium; PGC Consortium. An excess of risk-increasing low-frequency variants can be a signal of polygenic inheritance in complex diseases. Am J Hum Genet. 2014;94:437–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andreassen OA, Thompson WK, Schork AJ, et al. ; Psychiatric Genomics Consortium (PGC); Bipolar Disorder and Schizophrenia Working Groups. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holland D, Wang Y, Thompson WK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Enhancing Neuro Imaging Genetics through Meta Analysis Consortium. Estimating effect sizes and expected replication probabilities from GWAS summary statistics. Front Genet. 2016;7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maier R, Moser G, Chen GB, et al. ; Cross-Disorder Working Group of the Psychiatric Genomics Consortium. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am J Hum Genet. 2015;96:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Thompson WK, Schork AJ, et al. Leveraging genomic annotations and pleiotropic enrichment for improved replication rates in schizophrenia GWAS. PLoS Genet. 2016;12:e1005803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vilhjálmsson BJ, Yang J, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingason A, Giegling I, Hartmann AM, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC). Expression analysis in a rat psychosis model identifies novel candidate genes validated in a large case-control sample of schizophrenia. Transl Psychiatry. 2015;5:e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hargreaves A, Dublin TC, Anney R, et al. The one and the many: effects of the cell adhesion molecule pathway on neuropsychological function in psychosis. Psychol Med. 2017;44:2177–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sekar A, Bialas AR, de Rivera H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franke B, Stein JL, Ripke S, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; ENIGMA Consortium. Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci. 2016;19:420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee SH, Ripke S, Neale BM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruderfer DM, Fanous AH, Ripke S, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium; Cross-Disorder Working Group of the Psychiatric Genomics Consortium. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014;19:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee SH, Byrne EM, Hultman CM, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium and Rheumatoid Arthritis Consortium International; Schizophrenia Working Group of the Psychiatric Genomics Consortium Authors; Schizophrenia Working Group of the Psychiatric Genomics Consortium Collaborators; Rheumatoid Arthritis Consortium International Authors; Rheumatoid Arthritis Consortium International Collaborators. New data and an old puzzle: the negative association between schizophrenia and rheumatoid arthritis. Int J Epidemiol. 2015;44:1706–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levinson DF, Shi J, Wang K, et al. ; Schizophrenia Psychiatric GWAS Consortium. Genome-wide association study of multiplex schizophrenia pedigrees. Am J Psychiatry. 2012;169:963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamshere ML, Walters JT, Smith R, et al. ; Schizophrenia Psychiatric Genome-wide Association Study Consortium; Wellcome Trust Case Control Consortium+; Wellcome Trust Case Control Consortium 2. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the schizophrenia PGC. Mol Psychiatry. 2013;18:708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pers TH, Timshel P, Ripke S, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Comprehensive analysis of schizophrenia-associated loci highlights ion channel pathways and biologically plausible candidate causal genes. Hum Mol Genet. 2016;25:1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loh PR, Bhatia G, Gusev A, et al. ; Schizophrenia Working Group of Psychiatric Genomics Consortium. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47:1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee SH, DeCandia TR, Ripke S, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ripke S, Neale BM, Corvin A, et al. Consortium SWGotPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ripke S, Sanders AR, Kendler KS, et al. Consortium TSPG-WASG. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lencz T, Malhotra AK. Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol Psychiatry. 2015;20:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kotov R, Ruggero CJ, Krueger RF, Watson D, Yuan Q, Zimmerman M. New dimensions in the quantitative classification of mental illness. Arch Gen Psychiatry. 2011;68:1003–1011. [DOI] [PubMed] [Google Scholar]
- 31. Insel T, Cuthbert B, Garvey M, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. [DOI] [PubMed] [Google Scholar]
- 32. Cuthbert BN. Research domain criteria: toward future psychiatric nosologies. Dialogues Clin Neurosci. 2015;17:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cuthbert BN. Translating intermediate phenotypes to psychopathology: the NIMH research domain criteria. Psychophysiology. 2014;51:1205–1206. [DOI] [PubMed] [Google Scholar]
- 34. Cuthbert BN. The RDoC framework: facilitating transition from ICD/DSM to dimensional approaches that integrate neuroscience and psychopathology. World Psychiatry. 2014;13:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cuthbert BN; NIMH RDoC Workgroup The RDoC framework: continuing commentary. World Psychiatry. 2014;13:196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuthbert BN. Response to Lilienfield. Behav Res Ther. 2014;62:140–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cuthbert BN, Kozak MJ. Constructing constructs for psychopathology: the NIMH research domain criteria. J Abnorm Psychol. 2013;122:928–937. [DOI] [PubMed] [Google Scholar]
- 39. Fiorillo A, Luciano M, Del Vecchio V, Sampogna G, Obradors-Tarragó C, Maj M; ROAMER Consortium Priorities for mental health research in Europe: a survey among national stakeholders’ associations within the ROAMER project. World Psychiatry. 2013;12:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Os J, van der Steen Y, Islam MA, Gülöksüz S, Rutten BP, Simons CJ; GROUP Investigators Evidence that polygenic risk for psychotic disorder is expressed in the domain of neurodevelopment, emotion regulation and attribution of salience. Psychol Med. 2017;47:2421–2437. [DOI] [PubMed] [Google Scholar]
- 41. Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Os J. ‘Salience syndrome’ replaces ‘schizophrenia’ in DSM-V and ICD-11: psychiatry’s evidence-based entry into the 21st century?Acta Psychiatr Scand. 2009;120:363–372. [DOI] [PubMed] [Google Scholar]
- 43. Meehl PE. Schizotaxia, schizotypy, schizophrenia. Am Psychol. 1962;17:827–838. [Google Scholar]
- 44. Debbané M, Mohr C. Integration and development in schizotypy research: an introduction to the special supplement. Schizophr Bull. 2015;41(suppl 2):S363–S365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ettinger U, Mohr C, Gooding DC, et al. Cognition and brain function in schizotypy: a selective review. Schizophr Bull. 2015;41(suppl 2):S417–S426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cohen AS, Mohr C, Ettinger U, Chan RC, Park S. Schizotypy as an organizing framework for social and affective sciences. Schizophr Bull. 2015;41(suppl 2):S427–S435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cohen AS, Callaway DA, Najolia GM, Larsen JT, Strauss GP. On “risk” and reward: investigating state anhedonia in psychometrically defined schizotypy and schizophrenia. J Abnorm Psychol. 2012;121:407–415. [DOI] [PubMed] [Google Scholar]
- 48. Morton SE, O’Hare KJM, Maha JLK, et al. Testing the validity of taxonic schizotypy using genetic and environmental risk variables. Schizophr Bull. 2017;43:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Linscott RJ, Morton SE. The latent taxonicity of schizotypy in biological siblings of probands with schizophrenia. Schizophr Bull. 2017. doi:10.1093/schbul/sbx143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Linscott RJ, van Os J. Systematic reviews of categorical versus continuum models in psychosis: evidence for discontinuous subpopulations underlying a psychometric continuum. Implications for DSM-V, DSM-VI, and DSM-VII. Annu Rev Clin Psychol. 2010;6:391–419. [DOI] [PubMed] [Google Scholar]
- 51. Kerns JG, Nuechterlein KH, Braver TS, Barch DM. Executive functioning component mechanisms and schizophrenia. Biol Psychiatry. 2008;64:26–33. [DOI] [PubMed] [Google Scholar]
- 52. Kerns JG, Cohen JD, MacDonald AW III, Cho RY, Stenger VA, Carter CS. Anterior cingulate conflict monitoring and adjustments in control. Science. 2004;303:1023–1026. [DOI] [PubMed] [Google Scholar]
- 53. Cicero DC, Docherty AR, Becker TM, Martin EA, Kerns JG. Aberrant salience, self-concept clarity, and interview-rated psychotic-like experiences. J Pers Disord. 2015;29:79–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karcher NR, Martin EA, Kerns JG. Examining associations between psychosis risk, social anhedonia, and performance of striatum-related behavioral tasks. J Abnorm Psychol. 2015;124:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Berenbaum H, Valera EM, Kerns JG. Psychological trauma and schizotypal symptoms. Schizophr Bull. 2003;29:143–152. [DOI] [PubMed] [Google Scholar]
- 56. Kendler KS. The clinical features of paranoia in the 20th century and their representation in diagnostic criteria from DSM-III through DSM-5. Schizophr Bull. 2017;43:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grant P, Green MJ, Mason OJ. Models of schizotypy: the importance of conceptual clarity. Schizophr Bull. 2018. doi:10.1093/schbul/sby012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Debbané M, Eliez S, Badoud D, Conus P, Flückiger R, Schultze-Lutter F. Developing psychosis and its risk states through the lens of schizotypy. Schizophr Bull. 2015;41(suppl 2):S396–S407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fusar-Poli P, Borgwardt S, Bechdolf A, et al. The psychosis high-risk state: a comprehensive state-of-the-art review. JAMA Psychiatry. 2013;70:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lenzenweger MF. Schizotypy, schizotypic psychopathology and schizophrenia. World Psychiatry. 2018;17:25–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lord C, Veenstra-VanderWeele J. Following the trail from genotype to phenotypes. JAMA Psychiatry. 2016;73:7–8. [DOI] [PubMed] [Google Scholar]
- 62. Weiner DJ, Wigdor EM, Ripke S, et al. ; iPSYCH-Broad Autism Group; Psychiatric Genomics Consortium Autism Group. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. 2017;49:978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. St Pourcain B, Robinson EB, Anttila V, et al. ; iPSYCH-SSI-Broad Autism Group. ASD and schizophrenia show distinct developmental profiles in common genetic overlap with population-based social communication difficulties. Mol Psychiatry. 2018;23:263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ayalew M, Le-Niculescu H, Levey DF, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McLaughlin RL, Schijven D, van Rheenen W, et al. ; Project MinE GWAS Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun. 2017;8:14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nelson MT, Seal ML, Pantelis C, Phillips LJ. Evidence of a dimensional relationship between schizotypy and schizophrenia: a systematic review. Neurosci Biobehav Rev. 2013;37:317–327. [DOI] [PubMed] [Google Scholar]
- 68. Bigdeli TB, Bacanu SA, Webb BT, et al. Molecular validation of the schizophrenia spectrum. Schizophr Bull. 2014;40:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cannon TD, van Erp TG, Glahn DC. Elucidating continuities and discontinuities between schizotypy and schizophrenia in the nervous system. Schizophr Res. 2002;54:151–156. [DOI] [PubMed] [Google Scholar]
- 70. Gur RE, Calkins ME, Gur RC, et al. The consortium on the genetics of schizophrenia: neurocognitive endophenotypes. Schizophr Bull. 2007;33:49–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cicero DC, Kerns JG. Multidimensional factor structure of positive schizotypy. J Pers Disord. 2010;24:327–343. [DOI] [PubMed] [Google Scholar]
- 72. Tarbox SI, Pogue-Geile MF. A multivariate perspective on schizotypy and familial association with schizophrenia: a review. Clin Psychol Rev. 2011;31:1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kwapil TR, Barrantes-Vidal N. Schizotypy: looking back and moving forward. Schizophr Bull. 2015;41(suppl 2):S366–S373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fonseca-Pedrero E, Ortuño-Sierra J, de Álbeniz AP, Muñiz J, Cohen AS. A latent profile analysis of schizotypal dimensions: associations with psychopathology and personality. Psychiatry Res. 2017;253:110–115. [DOI] [PubMed] [Google Scholar]
- 75. Cohen AS, Docherty NM. Deficit versus negative syndrome in schizophrenia: prediction of attentional impairment. Schizophr Bull. 2004;30:827–835. [DOI] [PubMed] [Google Scholar]
- 76. Cohen AS, Dinzeo TJ, Nienow TM, Smith DA, Singer B, Docherty NM. Diminished emotionality and social functioning in schizophrenia. J Nerv Ment Dis. 2005;193:796–802. [DOI] [PubMed] [Google Scholar]
- 77. Cohen AS, Docherty NM. Symptom-oriented versus syndrome approaches to resolving heterogeneity of neuropsychological functioning in schizophrenia. J Neuropsychiatry Clin Neurosci. 2005;17:384–390. [DOI] [PubMed] [Google Scholar]
- 78. Cohen AS, Alpert M, Nienow TM, Dinzeo TJ, Docherty NM. Computerized measurement of negative symptoms in schizophrenia. J Psychiatr Res. 2008;42:827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cohen AS, Nienow TM, Dinzeo TJ, Docherty NM. Attribution biases in schizophrenia: relationship to clinical and functional impairments. Psychopathology. 2009;42:40–46. [DOI] [PubMed] [Google Scholar]
- 80. Kendler KS, McGuire M, Gruenberg AM, Walsh D. Schizotypal symptoms and signs in the Roscommon Family Study. Their factor structure and familial relationship with psychotic and affective disorders. Arch Gen Psychiatry. 1995;52:296–303. [DOI] [PubMed] [Google Scholar]
- 81. Chan RC, Gooding DC, Shi HS, et al. Evidence of structural invariance across three groups of Meehlian schizotypes. NPJ Schizophr. 2016;2:16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kendler KS, Neale MC. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15:789–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fonseca-Pedrero E, Paíno M, Lemos-Giráldez S, et al. Schizotypy assessment: state of the art and future prospects. Int J Clin Health Psychol. 2017;8:577–593. [Google Scholar]
- 84. Debbané M, Barrantes-Vidal N. Schizotypy from a developmental perspective. Schizophr Bull. 2015;41(suppl 2):S386–S395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Docherty AR, Sponheim SR.. Anhedonia as an Indicator of Genetic Vulnerability to Schizophrenia. Vol 2 Dordrecht: Springer; 2014:105–123. [Google Scholar]
- 86. Chan RC, Di X, McAlonan GM, Gong QY. Brain anatomical abnormalities in high-risk individuals, first-episode, and chronic schizophrenia: an activation likelihood estimation meta-analysis of illness progression. Schizophr Bull. 2011;37:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Grazioplene RG, Chavez RS, Rustichini A, DeYoung CG. White matter correlates of psychosis-linked traits support continuity between personality and psychopathology. J Abnorm Psychol. 2016;125:1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hengartner MP, Lehmann SN. Why psychiatric research must abandon traditional diagnostic classification and adopt a fully dimensional scope: two solutions to a persistent problem. Front Psychiatry. 2017;8:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kotov R, Chang SW, Fochtmann LJ, et al. Schizophrenia in the internalizing-externalizing framework: a third dimension?Schizophr Bull. 2011;37:1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Keyes KM, Eaton NR, Krueger RF, et al. Thought disorder in the meta-structure of psychopathology. Psychol Med. 2013;43:1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kraepelin E, Defendorf AR.. Clinical Psychiatry: A Text-Book for Students and Physicians: Abstracted and Adapted from the 6th German Edition of Kraepelin’s “Lehrbuch der Psychiatrie.” London: Macmillan; 1904. [Google Scholar]
- 92. Berrios GE, Beer D. The notion of a unitary psychosis: a conceptual history. Hist Psychiatry. 1994;5:13–36. [DOI] [PubMed] [Google Scholar]
- 93. Carpenter WT, Strauss JS. Developmental interactive framework for psychotic disorders. Schizophr Bull. 2017;43:1143–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Stefanovics EA, Krystal JH, Rosenheck RA. Symptom structure and severity: a comparison of responses to the Positive and Negative Syndrome Scale (PANSS) between patients with PTSD or schizophrenia. Compr Psychiatry. 2014;55:887–895. [DOI] [PubMed] [Google Scholar]
- 95. Emsley R, Rabinowitz J, Torreman M; RIS-INT-35 Early Psychosis Global Working Group The factor structure for the Positive and Negative Syndrome Scale (PANSS) in recent-onset psychosis. Schizophr Res. 2003;61:47–57. [DOI] [PubMed] [Google Scholar]
- 96. Wigman JT, Vollebergh WA, Jacobs N, et al. Replication of the five-dimensional structure of positive psychotic experiences in young adulthood. Psychiatry Res. 2012;197:353–355. [DOI] [PubMed] [Google Scholar]
- 97. Shevlin M, McElroy E, Bentall RP, Reininghaus U, Murphy J. The psychosis continuum: testing a bifactor model of psychosis in a general population sample. Schizophr Bull. 2017;43:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Reininghaus U, Böhnke JR, Hosang G, et al. Evaluation of the validity and utility of a transdiagnostic psychosis dimension encompassing schizophrenia and bipolar disorder. Br J Psychiatry. 2016;209:107–113. [DOI] [PubMed] [Google Scholar]
- 99. McGorry PD, Nelson B, Markulev C, et al. Effect of ω-3 polyunsaturated fatty acids in young people at ultrahigh risk for psychotic disorders: the NEURAPRO randomized clinical trial. JAMA Psychiatry. 2017;74:19–27. [DOI] [PubMed] [Google Scholar]
- 100. Schultze-Lutter F, Michel C, Schmidt SJ, et al. EPA guidance on the early detection of clinical high risk states of psychoses. Eur Psychiatry. 2015;30:405–416. [DOI] [PubMed] [Google Scholar]
- 101. Fusar-Poli P, Carpenter WT, Woods SW, McGlashan TH. Attenuated psychosis syndrome: ready for DSM-5.1?Annu Rev Clin Psychol. 2014;10:155–192. [DOI] [PubMed] [Google Scholar]
- 102. Tarbox SI, Almasy L, Gur RE, Nimgaonkar VL, Pogue-Geile MF. The nature of schizotypy among multigenerational multiplex schizophrenia families. J Abnorm Psychol. 2012;121:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Macare C, Bates TC, Heath AC, Martin NG, Ettinger U. Substantial genetic overlap between schizotypy and neuroticism: a twin study. Behav Genet. 2012;42:732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jones HJ, Stergiakouli E, Tansey KE, et al. Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry. 2016;73:221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fanous AH, Zhou B, Aggen SH, et al. ; Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study of clinical dimensions of schizophrenia: polygenic effect on disorganized symptoms. Am J Psychiatry. 2012;169:1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]