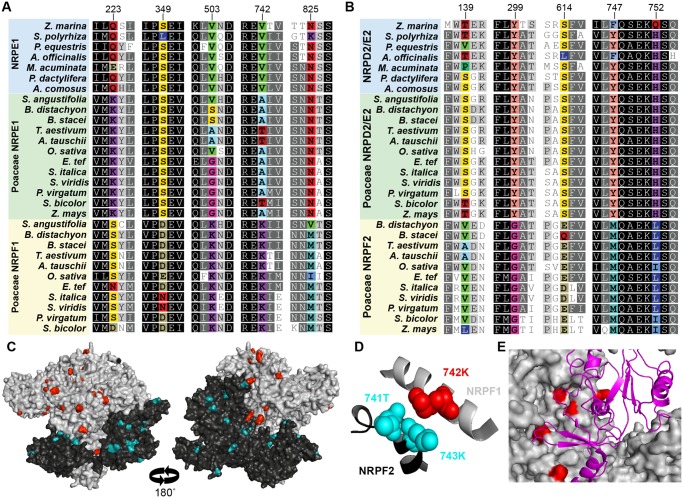
Fig. 3.
Sites under positive selection cluster on the surface of Pol VI subunits. Alignment of monocot largest (A) and second largest (B) subunits illustrates a subset of the residues that are under positive selection following gene duplication (colored). Remaining residues are colored based on sequence conservation. Numbers at top are the amino acid position in the Oryza sativa NRPF1 and NRPF2 and correspond with supplementary tables 4 and 5, Supplementary Material online. (C) Residues under positive selection (colored) are found on the surface of homology-based structures of NRPF1 (gray) and NRPF2 (black). (D) Residues under positive selection also occur at the interface between subunits, as demonstrated by 742K in NRPF1, which is directly opposite 741T and 743K in NRPF2. (E) Several residues under positive selection are found where the largest subunit (gray) interacts with the fifth subunit (magenta ribbon).