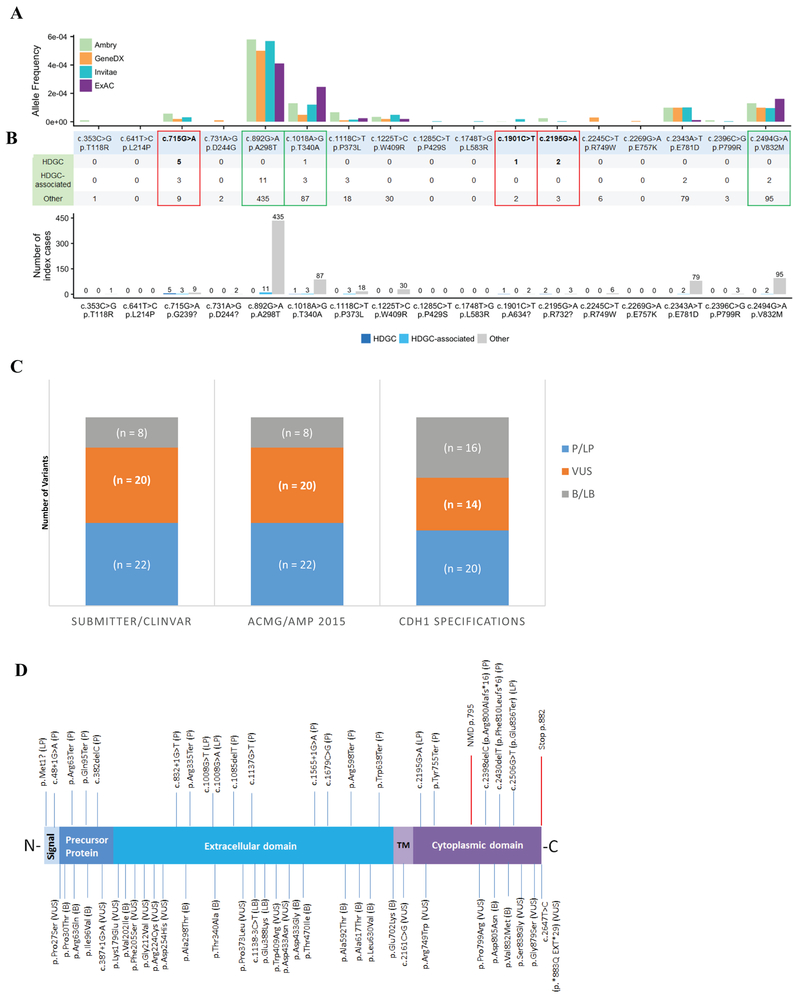
Figure 1 –
Pilot assessment of CDH1 variants.
A – Clinical and population evidence of selected CDH1 missense variants. Red boxes highlight the three missense variants that were demonstrated functionally by RNA analysis to activate cryptic splicing sites; green boxes highlight variants that met benign population allele-frequency cutoffs. Allele frequencies for the selected variants in the ExAC database (~120,000 alleles) and in the cohorts of the three largest data sharing clinical genetic laboratories in the U.S.A. (~827,000 alleles total).
B – Laboratory cohort phenotypes associated with each of the specific variants. HDGC = family meets International Gastric Cancer Linkage Consortium (IGCLC) criteria; HDGC-associated tumors = Diffuse Gastric Cancer, Signet Ring Cell tumors (e.g., Krukenberg tumor), Lobular Breast Cancer, but does not meet IGCLC criteria; Others are either reported unaffected or having non-CDH1 tumors (e.g., breast invasive ductal carcinoma).
C – Comparing variant classifications from EP or ClinVar submitters and from the original ACMG/AMP criteria versus applying CDH1 evidence code specifications demonstrates a reduction of VUS. P = pathogenic; LP = likely pathogenic; VUS = variant of uncertain significance; LB = likely benign; B = benign. ClinVar assertions were accessed on April 16, 2018.
D – E-cadherin schematic representation demonstrating its major domains in relation to the 50 pilot variants curated by the EP. E-cadherin is a highly conserved transmembrane protein that possess a signal peptide, a precursor protein, the extracellular domain, which is the “adhesive” ectodomain composed of tandem cadherin repeats, the transmembrane (TM) domain, and the cytoplasmic domain, which interacts with the armadillo catenins, p120 and β-catenin (Gul et al., 2017). Clinically actionable variants (P, LP) are shown at the top; VUS, LB, and B variants are shown at the bottom.