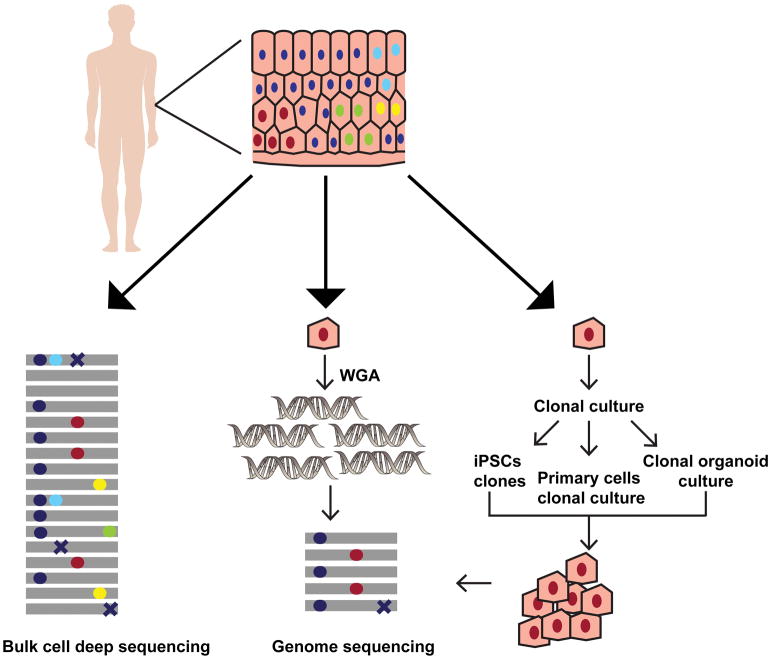
Figure 1. Overview of the methods for detecting somatic mutations in healthy human cells.
The various approaches to analyze somatic mutations in a heterogenous tissue sample are depicted using skin as an example. The colored circles (red, cyan, yellow and green) in the tissue denote nuclei within the cells with different somatic mutations respectively. The dark blue circles denote nuclei with germline polymorphisms present across all cells in the tissue. DNA extraction from the bulk cells followed by deep sequencing provides a measure of the diverse somatic mutations present in the sample (left panel). The gray lines denote individual reads in NGS, and the colored circles on them imply heterozygous somatic mutations or germline polymorphisms present in the tissue. The somatic mutations in bulk sequencing samples are often present in very low allele fractions. In addition, sequencing errors (crosses) may further confound analysis in deep-sequenced heterogenous samples. To determine the somatic mutations in a given cell whole genome amplification from a single cell followed by WGS may be used (middle panel). On the other hand, pluripotent stem cells may be derived from the tissue and grown clonally to get enough number of cells for WGS. The primary cells may also be either cultured directly, or organoid cultures may be established from the stem cells within the tissue allowing propagation in vitro to obtain DNA for WGS (right panel). In single cell sequencing, and clonal population sequencing somatic mutations are present as high frequency alleles, while errors generated during library preparation and sequencing are usually present in a smaller fraction of reads. Therefore, somatic mutation calling using these approaches is more accurate.