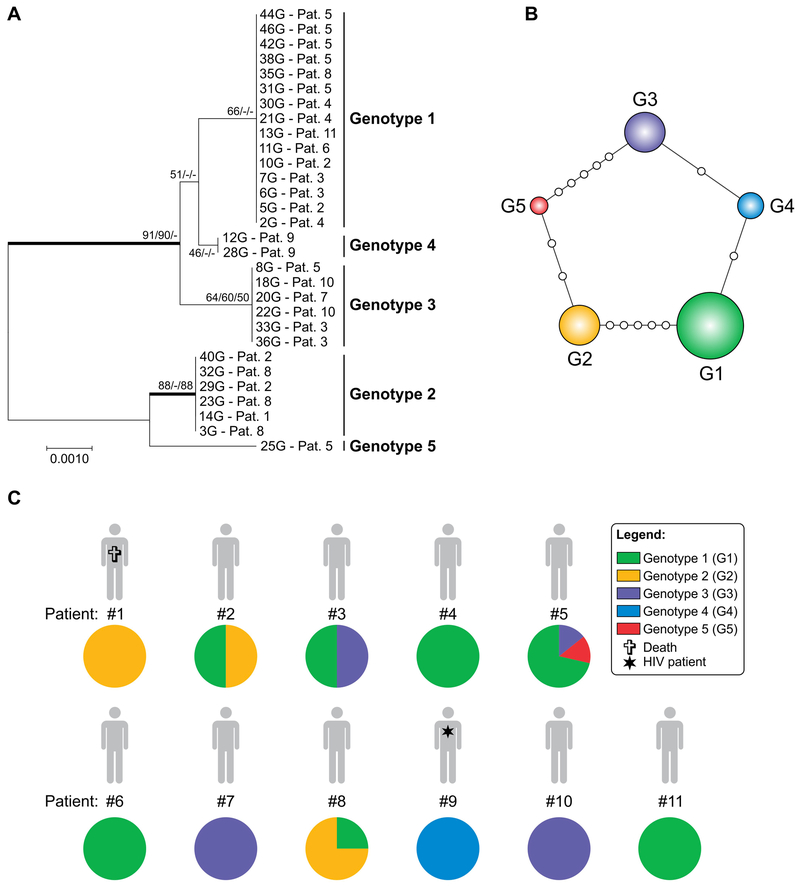
Figure 1.
Genetic diversity of Pneumocystis jirovecii pneumonia during a cluster in renal transplant recipients (RTRs). A, Phylogenetic relationships, as inferred from a neighbour‐joining analysis (model T92) of the concatenated nuclear 26S and the mitochondrial 23S mtLSU sequences from 28 samples obtained from 10 RTRs and 2 samples from one (1) HIV‐infected patient (pt. 9). The numbers close to the branches represent indices of support (NJ/ML/MP) based on 1000 bootstrap replications. The branches with bootstrap support higher than 80% are indicated in bold. NJ, neighbour‐joining; ML, maximum likelihood; MP, maximum parsimony. B, Median‐joining haplotype/genotype network of P. jirovecii samples, covering all genotypes (G_1–G_5) found in this study. The size of the circumference is proportional to the genotype frequency in the dataset. Mutational steps are represented by white dots. C, The genotypes are colour coded, and their frequencies per patient are shown. Coinfection with at least two different P. jirovecii genotypes was found in four of the eleven patients evaluated. Further information about isolate source can be found in the Table S2