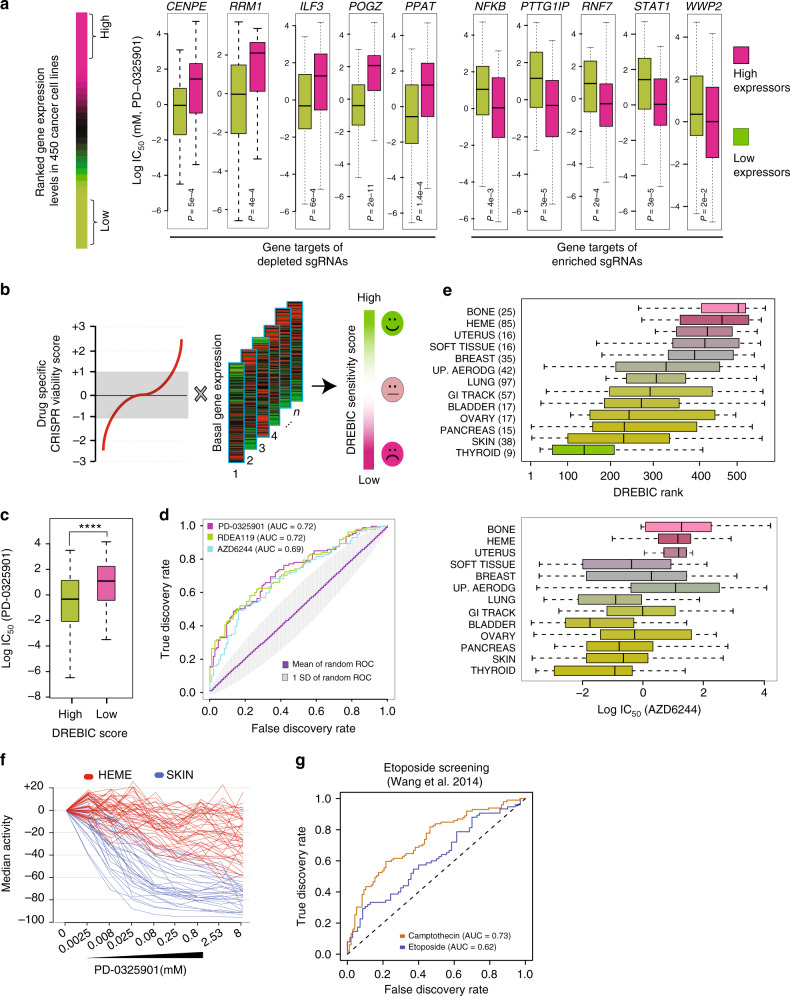
Fig. 4.
Drug response evaluation by in vivo CRISPR Screening (DREBIC) approach. a The 450 CGP cancer cell lines15 were ranked according to the expression levels of the indicated genes. Box plots depict the distribution of log IC50 growth inhibition values of MEK inhibitor PD-0325901 in the high- (top quartile) and low-expressing cell lines (bottom quartile). Statistical significance was calculated by Kolmogorov–Smirnov test. Comparable results were obtained for other MEK inhibitors. b DREBIC integrates gene-specific CRISPR viability scores with basal expression levels to model drug response phenotype. c Box plot showing a significant (p < 0.0001, Kolmogorov–Smirnov test) growth inhibition (IC50) difference in response to PD-0325901 MEK inhibitor in ~450 cancer cell lines scoring high (top quartile) or low (bottom quartile) in DREBIC score analysis. d Receiver operation characteristics curves demonstrating true vs. false discovery rate of DREBIC prediction of cellular responses to three separate MEK inhibitors (PD-0325901 red, AZD6244 blue, and RDEA119 green). The random DREBIC score distribution was generated by 10,000 permutations of the same number of genes (Supplementary Figure 12). The average ROC curve of permutation analysis is shown in purple (AUC = 0.5) and the area corresponding to one standard deviation of prediction is marked in gray. e The box plots in the top panel are showing the distribution of tissue-specific DREBIC scores of CGP cell lines as ordered by ascending median DREBIC score. The lower panel box plots are showing log IC50 growth inhibition values of AZD6244 MEK inhibitor drug for the same cancer types. f The PD-0325901 MEK inhibitor drug response curves are shown for hematopoietic (Heme, red) and melanoma cancer cell lines (Skin, blue) from the CCLE data set13. g The ROC curve shows the accuracy rate of etoposide-based DREBIC analysis of cellular response to three independent topoisomerase inhibitors from CPG data set. The CRISPR-gene viability scores for etoposide were obtained from a previously published in vitro CRISPR screening by Wang et al.11. In the box plots, bounds of the box spans from 25 to 75% percentile, center line represents median, and whiskers visualize 5 and 95% of the data points. Symbol **** denotes P < 0.0001