Fig. 2.
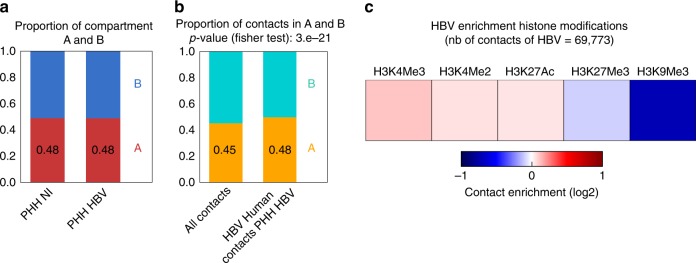
HBV genome contacts are enriched at active chromatin. a Distribution of active A and inactive B compartments in non-infected and infected PHH (400 kb resolution). b Distribution of Hi-C reads coverage in compartments A and B and distribution of HBV reads contacts on the genome in infected PHH (400 kb resolution) (statistical analysis was performed using Fisher's test). c Heatmap of HBV contacts enrichment for different histone modification marks (computed in windows ± 3.5 kb around the start of the read; N realizations = 100). Signals for the heatmap are represented by fold change (log2 ratio) compared to the average of random groups (see Methods). Histone modification marks dataset were from the Encode project generated using hepatic HepG2 cells (Encode Project:GSE29611)15