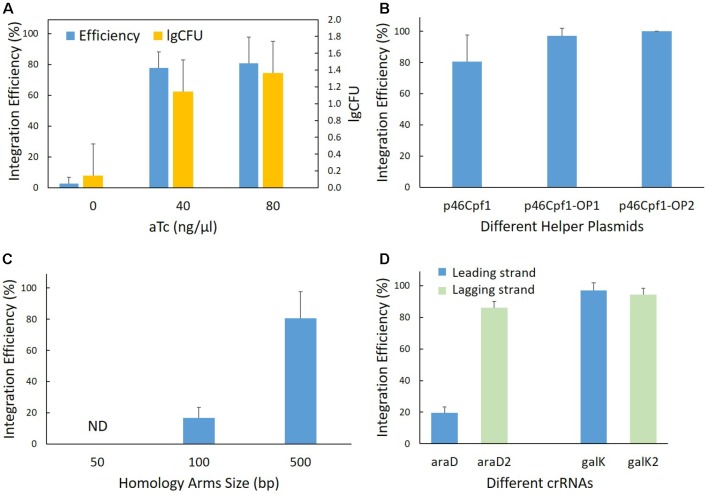
FIGURE 3.
Parameters affecting recombination efficiency. (A) Effect of the λ-Red machinery. Genomic integration was performed at the pyrF site in both the λ-Red induced (aTc added) and non-induced (aTc not added) backgrounds. The helper plasmid p46Cpf1 and the donor plasmid pTc-PG were used. CFU stands for colony number of forming units per microliter of culture. (B) Effect of codon optimization of the Cas12a gene. Genomic integration was performed at the pyrF site using different helper plasmids - p46Cpf1 with the original Cas12a (Cpf1) gene, p46Cpf1-OP1 and p46Cpf1-OP2 with codon optimized Cas12a genes. The donor plasmid pTc-PG was used. (C) Effect of homology arm length on integration efficiency. Genomic integration was performed at the pyrF site with different donor plasmids, pTc-P-50bp providing the donor template with 50-bp homology arms, pTc-P-100bp with 100-bp homology arms and pTc-P with 500-bp homology arms. The helper plasmid p46Cpf1 was used. Modified colonies were not detected when using 50 bp homology arms. ND, not detected. (D) Relationship between integration efficiency and different crRNAs. The crRNAs araD and galK target the leading strand in the E. coli genome, while the crRNAs araD2 and galK2 target the lagging strand. The helper plasmid p46Cpf1 was used. The data represent the averages of three independent experiments.