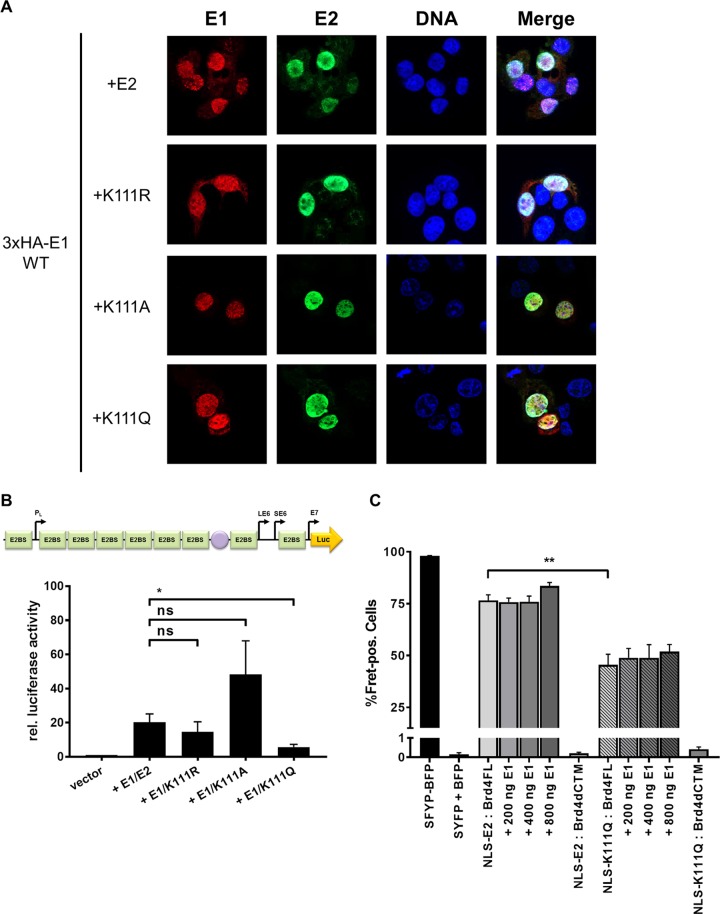
FIG 6.
E2 K111 mutant proteins relocalize to nucleus in the presence of CRPV E1. (A) C33a cells were cotransfected with 500 ng E1 expression vector (pSG 3×HA:CRPV E1) and 500 ng E2 expression vectors (as indicated on the left). Forty-eight hours after transfection, cells were stained with an anti-HA antibody to detect the E1 protein (red) and an anti-CRPV E2 antibody to detect E2 proteins (green) and analyzed by immunofluorescence microscopy. DNA was stained with DAPI (blue). (B) C33a cells were transiently transfected with 50 ng pGL CRPV (PL-PE7) firefly reporter construct, 100 ng E1 expression vector (pSG CRPV E1), and 10 ng of the different E2 expression vectors. Empty expression vector (pSG5) was used to adjust differences in DNA amounts. The values represent the relative luciferase activities of E1/E2 (E2 wild-type or E2 K111 mutant proteins) to the basal activity of the used firefly reporter construct. Error bars indicate the standard errors of the means (SEM) from at least five independent experiments. A paired two-tailed t test was used to determine statistical significance (*, P < 0.05). ns, nonsignificant. (C) C33a cells were cotransfected with expression vectors for BFP and SYFP fusion proteins and, as indicated, increasing DNA amounts of CRPV E1 expression vector (pSG CRPV E1). SYFP-BFP fusion protein was used as a positive control, and cotransfected BFP and SYFP expression vectors were negative controls. Shown are the mean values of FRET-positive cells from at least three independent experiments. Error bars indicate the SEM. A paired two-tailed t test was used to determine statistical significance (**, P < 0.01).