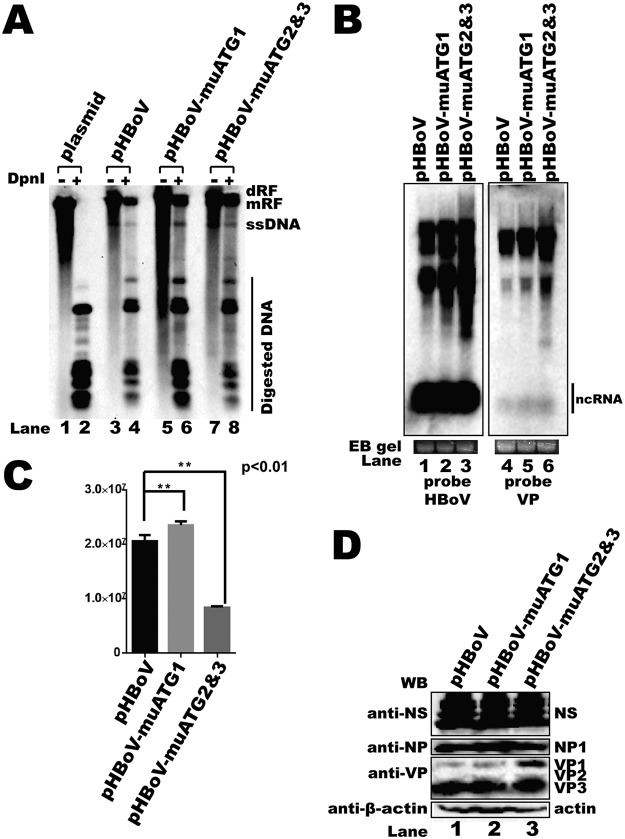
FIG 8.
uATG2 and uATG3 knockout mutations in an HBoV1 infectious clone decreased viral progeny production. (A) Southern blot. Hirt DNA was isolated from HEK293T cells transfected with plasmids, as indicated, at 48 h posttransfection. The DpnI-digested fragments were resolved on 1% agarose gels and transferred to a Hybond-N+ membrane, followed by hybridization with an HBoV1 probe spanning from nt 1 to 5543. dRF, double replication form; mRF, monomer replication form; ssDNA, single-stranded DNA. (B) Northern blot. Total RNAs were harvested from transfected cells, and Northern blotting was carried out as described in the legend to Fig. 2D. Ethidium bromide (EB)-stained 18S RNA bands are shown as the loading control. The images in panel B were taken from different areas of the same photo or different gels and then joined together. (C) Quantitative PCR (qPCR) for the HBoV1 genome. Cell lysates of uATG-mutated HBoV1 infectious clones were prepared by four cycles of freezing and thawing, followed by DNase I digestion. The progeny viruses were separated by cesium chloride (CsCl) density gradient centrifugation, and genomic DNA was prepared as the template for qPCR. Means and standard deviations were calculated from the results of at least three independent experiments. The pHBoV plasmid was used as a control to establish a standard curve for absolute qPCR (E = 105.0%, R2 = 0.999, slope = −3.208). E, efficiency. (D) Western blot. uATG-mutated infectious clones were transfected into HEK293T cells, and Western blotting was performed with anti-NS, anti-NP, and anti-VP antibodies.