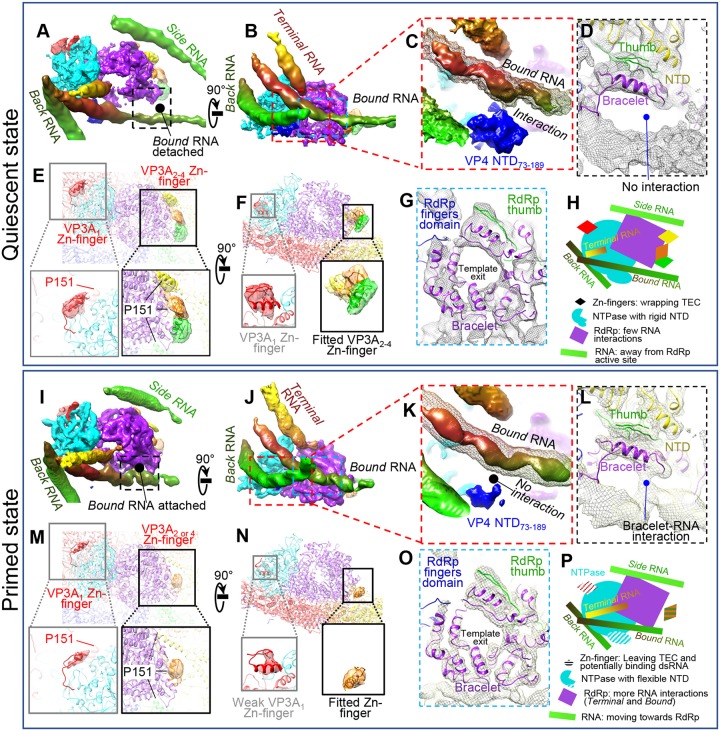
FIG 14.
Comparison between quiescent and primed ARV shows similarities and differences in RNA, TEC, and CSP. (A to H) Structural details of a TEC under a polar vertex in the quiescent state. (A and B) Two orthogonal views showing four RNAs interacting with TECs. (C) Enlargement of the area boxed in red in panel B showing interaction of NTPase NTD73–189 with bound RNA (mesh). The solid density is at a higher display threshold than the mesh. (D) Enlargement of the boxed area in panel A showing detachment of the bound RNA from the RdRp in the quiescent state. (E and F) Magnified vertices showing only CSP N-terminal densities, with four structured densities (VP3A1 to VP3A4 Zn fingers, aa 117 to 141) labeled. P151 locations in VP3A1 to VP3A4 conformers are labeled to show the close proximity between N anchors and the corresponding Zn fingers. (G) Enlargement of the area boxed in blue in panel B showing the bracelet domain. (H) Cartoon of the relative positions of RNA and TEC. (I to P) Same view as in panels A to H for primed-state structures. Key differences from the quiescent state are as follows: bound RNA attaches to the RdRp bracelet domain (A, D, G, and L), the NTD73–189 domain in NTPase becomes more flexible (C and K), and the VP3A Zn finger becomes more flexible (E, F, M, and N). Unlike CPV, no large conformation changes in the bracelet domain could be found (G and O). (P) Cartoon of TEC and RNA structures in the primed state. All the map segmentations are based on the Chimera map segmentation function. No atomic model was used to generate the segmentation.