Abstract
After liver injury, regeneration manifests as either (1) hepatocytes proliferating to restore the lost hepatocyte mass or (2) if hepatocyte proliferation is compromised, biliary epithelial cells (BECs) dedifferentiating into liver progenitor cells (LPCs), which subsequently differentiate into hepatocytes. Following pharmacogenetic ablation of hepatocytes in Tg(fabp10a:CFP-NTR) zebrafish, resulting in severe liver injury, signal transducer and activator of transcription 3 (Stat3) and its target gene and negative regulator, socs3a, were upregulated in regenerating livers. Using either Stat3 inhibitors, JSI-124 and S3I-201, or stat3 zebrafish mutants, we investigated the role of Stat3 in LPC-driven liver regeneration. Although Stat3 suppression reduced the size of regenerating livers, BEC dedifferentiation into LPCs was unaffected. However, regenerating livers displayed a delay in LPC-to-hepatocyte differentiation and a significant reduction in the number of BECs. While no difference in cell death was detected, Stat3 inhibition significantly reduced LPC proliferation. Notably, stat3 mutants phenocopied the effects of Stat3 chemical inhibitors, although the mutant phenotype was incompletely penetrant. Intriguingly, a subset of socs3a mutants also displayed a lower number of BECs in regenerating livers. We conclude that the Stat3/Socs3a pathway is necessary for the proper timing of LPC-to-hepatocyte differentiation and establishing the proper number of BECs during LPC-driven liver regeneration.
Key words: Liver progenitor cells (LPCs), Biliary epithelial cells (BECs), Oval cells, Socs3
INTRODUCTION
With the story of an eagle picking at Prometheus’ liver daily, the Ancient Greeks first alluded to the remarkable ability of the liver to regenerate. To date, no true stem cell population has been identified in the liver. Instead, extensive studies have revealed two main modes of liver regeneration depending on the severity of liver injury: (1) hepatocyte driven and (2) biliary epithelial cell (BEC) driven. Typically following liver injury, hepatocytes proliferate to restore the lost liver mass1,2. However, if hepatocyte proliferation is blocked, then BECs undergo dedifferentiation into hepatoblast-like cells (HB-LCs), also termed liver progenitor cells (LPCs)3 or oval cells. LPCs are bipotent4, facultative resident stem progenitor cells that can differentiate into either hepatocytes or BECs to restore the lost liver mass4–6.
Moreover, LPCs observed in a zebrafish LPC-driven liver regeneration model7,8 share similar characteristics with BEC-derived oval cells or ductular reactions observed in both rodent liver injury models9 and human severe acute or chronic liver injury conditions10. Currently, chronic liver diseases, including cirrhosis, are the 12th leading cause of death in the US11. Although liver transplantation offers an effective therapy, more than 50% of patients on the transplant waiting list do not receive a donor liver transplant in time12. The presence of LPCs can be double edged. Following severe liver injury, the bipotent LPCs, activated via BEC dedifferentiation, are needed to replenish the functional cells (i.e., hepatocytes) in the liver13. However, the continued presence of LPCs can be detrimental. In one study, in patients presenting with chronic liver disease, such as alcoholic hepatitis, LPC markers were increased and correlated with short-term mortality and disease severity14. In a separate study, the number of LPCs, detected in various liver diseases including non-alcoholic fatty liver disease, correlates with disease severity15–17. Furthermore, hepatocellular carcinoma liver cells expressing OV6, an LPC marker, have been linked to tumor progression due to the invasive/tumorigenic properties of OV6+ cells18. Thus, the continued presence of LPCs is undesirable and can be detrimental to functional liver recovery; however, if the LPCs can be coaxed to differentiate into hepatocytes, then generating more LPCs can prove beneficial for the innate regenerative process. Because of the shortage of donor livers and the implications of LPCs in disease progression, an essential step in treating liver disease and augmenting the innate regenerative process is a better understanding of the underlying mediators and mechanisms of LPC-driven liver regeneration, including LPC activation, proliferation, and differentiation.
With that goal in mind, our lab7 and others8,19 have previously reported on a zebrafish model, in which following near-complete hepatocyte ablation, BECs extensively contribute to hepatocytes by giving rise to LPCs. This phenomenon is not zebrafish exclusive, however, as the BEC-to-hepatocyte-mediated regeneration was similarly observed in several mouse models in which hepatocyte proliferation is greatly compromised6,20.
A separate model of LPC-mediated repair in mice is the choline-deficient, ethionine-supplemented (CDE) diet; mice fed a CDE diet display an increased release of cytokines, such as interleukin-6 (IL-6), after liver injury21. Once IL-6 binds to its cognate receptor and coreceptor, gp130-associated Janus tyrosine kinases (JAKs) are autophosphorylated and subsequently activated. JAKs can then phosphorylate transcription factors, such as signal transducer and activator (STAT) 3, which dimerize and are able to translocate to the nucleus, inducing the expression of downstream target genes22,23.
Previous reports have considered the involvement of Stat3 in hepatocyte-driven regeneration24 and suggested its role in oval cell proliferation25. In the partial hepatectomy (PHx) model of murine liver injury, Stat3 deficiency led to an increased mortality rate and slightly decreased hepatocyte proliferation24. In the CDE diet model of liver injury, Stat3 expression was upregulated in the oval cell population with high proliferative potential25. In addition, mice with a hepatocyte-specific knockout of suppressor of cytokine signaling 3 (Socs3), a negative regulator of STAT326, display STAT3 hyperactivation after PHx, which results in enhanced proliferation during regeneration27. Given the known involvement of Stat3 in hepatocyte survival and proliferation after liver injury, we hypothesized that in zebrafish, Stat3, along with Socs3, may also regulate LPC-driven liver regeneration.
Here using the zebrafish hepatocyte ablation model for LPC-driven liver regeneration, we report on the roles of Stat3 in LPC proliferation and LPC-to-hepatocyte differentiation during the regeneration. We investigated the roles of Stat3 and Socs3a in LPC-driven liver regeneration by two methods: (1) utilizing a JAK-specific inhibitor as well as a Stat3 dimerization inhibitor to block Stat3 downstream signaling and (2) utilizing zebrafish stat3 and socs3a mutants generated by genome editing technology. Our findings show that Stat3 plays an important role in regulating LPC proliferation as well as LPC differentiation into hepatocytes during LPC-driven liver regeneration. Moreover, Stat3 and Socs3a are important for regulating the proper number of BECs during the regeneration process.
MATERIALS AND METHODS
Zebrafish Maintenance
Embryos and adult zebrafish (Danio rerio) were raised and maintained under standard laboratory conditions28 with protocols approved by the University of Pittsburgh Institutional Animal Care and Use Committee. We used stat3 stl27 29 and socs3a ihb31 mutants and the following transgenic lines: Tg(fabp10a:DsRed,ela3l:GFP)gz12 30 [referred to as Tg(fabp10a:DsRed)], Tg(EPV.Tp1-Mmu.Hbb:Venus-Mmu.Odc1) s940 31 [referred to as Tg(Tp1:VenusPEST)], Tg(EPV.Tp1-Mmu.Hbb:hist2h21-mCherry) s939 31 [referred to as Tg(Tp1:H2B-mCherry)], Tg(fabp10a:CFP-NTR) s931 32, and Tg(fabp10a:mAGFP-gmnn,cryaa:ECFP) pt608 33 [referred to as Tg(fabp10a:mAGFP-gmnn)].
Genotyping of the stat3 stl27 Mutant Line
The wild-type allele was amplified by PCR with a forward primer 5′-CCTCCACAGGACATGGAG-3′ and a common reverse primer 5′-ATATTGTGGAACCCTGACCAAAAAACAACATTTCCAATGCAGTCATACCTCCAGCACTC-3′. The mutant allele was separately amplified with a forward primer 5′-CATCCTCCACAGGACAGA-3′ and the common reverse primer. While the wild-type allele generated a 139-bp band, the mutant allele generated a 135-bp band.
Generation of the socs3a Mutant Line
The construction of sequence-specific transcription activator-like effector nuclease (TALEN) effector repeats recognizing two targeting sites on exon 2 of socs3a (left: ACCACTTCAAGACCT and right: GCACCAGCTGAAACT) was performed using the unit assembly method as previously described34. To generate capped mRNA containing DNA-binding TALEN repeats and the FokI endonuclease domain, the TALEN expression vectors were linearized with NotI and transcribed using a Sp6 mRNA kit (Message Machine Kit; AM1340; Ambion). Capped mRNA was diluted to 200 ng/μl and injected into wild-type embryos at the one-cell stage. A primer pair [socs3a, TACTCATCCTGACTTTCTC (forward) and CCAGTCTCAGCACGCAGT (reverse)] was used to amplify the target region; the PCR products were digested with StyI restriction enzyme. The remaining mutation-positive larvae were raised to F0 adulthood and outcrossed with wild-type adults. The genomic DNA of F1 larvae was subject to PCR amplification, StyI digestion, and Sanger sequencing. The F1 population containing socs3a heterozygous larvae were raised to adulthood and genotyped. The socs3a mutant line with a 61-bp deletion from 221 to 281 bp of the wild-type socs3a coding sequence was established and is available at the China Zebrafish Resource Center.
Hepatocyte Ablation and Chemical Inhibitor Treatments
To ablate hepatocytes, Tg(fabp10a:CFP-NTR) larvae were treated with 10 mM metronidazole (Mtz) in egg water supplemented with 0.2% dimethyl sulfoxide (DMSO) and 0.2 mM 1-phenyl-2-thiourea (PTU) as previously described35. For Stat3 inhibitor experiments, larvae were treated with 7 μM JSI-124 (also known as Cucurbitacin I; Cayman Chemical, Ann Arbor, MI, USA) or 200 μM S3I-201 (Sigma-Aldrich, St. Louis, MO, USA). Larvae were treated with JSI-124 or S3I-201 from ablation 0 h (A0h) or A18h to regeneration 24 h (R24h) or R48h.
Zebrafish Whole-Mount In Situ Hybridization (WISH), qPCR, and Immunostaining
Whole-mount in situ hybridization was performed as previously described36. socs3a forward primer (5′-AAGACCTTCAGCTCCAAGGTG-3′) and reverse primer (5′-GTGTGTCCGTTCACAGTCTTCCT-3′) were used to amplify the socs3a region for in situ probes; the stat3 plasmid used for this PCR was a gift from Ken Poss at Duke University. For qPCR analysis, 100 livers were dissected for each condition, and total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The SuperScript III First-Strand Synthesis SuperMix (Life Technologies, Grand Island, NY, USA) was used to synthesize cDNA from the extracted RNA. Using the Bio-Rad iQ5 PCR machine with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA), qPCR was performed as previously described37. eef1b2 was used for normalization. The primers used for qPCR analysis were eef1b2 (forward: 5′-CCCTCTCAGGCTGATATTGC-3′; reverse: 5′-TAAGCTGCAAGCCTCTCCTC-3′), socs3a (forward: 5′-CACTAACTTCTCTAAAGCAGGG-3′; reverse: 5′-GGTCTTGAAGTGGTAAAACG-3′), and stat3 (forward: 5′-GGACTTCCCGGACAGTGAG-3′; reverse: 5′-ATCGCTTGTGTTGCCAGAG-3′). Whole-mount immunostaining was performed as previously described32 using the following antibodies: goat anti-Hnf4a (1:70; Santa Cruz, Dallas, TX, USA), mouse monoclonal anti-Bhmt (1:400; a gift from J. Peng, Zhejiang University, P.R. China), mouse monoclonal anti-Alcam (1:10; ZIRC; Eugene, OR, USA), rabbit polyclonal anti-Abcb11 (1:500; PC-064; Kamiya Biomedical, Seattle, WA, USA), and Alexa Fluor 488-, 568-, and 647-conjugated secondary antibodies (1:300; Life Technologies).
TUNEL and EdU Assays
Apoptotic cell death was analyzed according to the protocol of the In Situ Cell Death Detection Kit, TMR Red (Roche, Switzerland). Following whole-mount immunostaining, terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) labeling was applied. Cell proliferation was performed using the protocol outlined in the Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technologies). Larvae were treated with egg water supplemented with 0.2 mM PTU containing 50 mM EdU and 5% DMSO at R0h or R18h. After a 6-h treatment window, larvae were harvested at R6h or R24h, respectively, and fixed in fresh 3% formaldehyde overnight at 4°C. After fixation, samples were prepared for subsequent imaging and statistical analysis.
BODIPY C5 Lipid Analog Assay
A 2-h treatment of 0.5 μM BODIPY C5 (Life Technologies)38 was used to assess ductal morphology and morphogenesis in regenerating livers. Following BODIPY C5 treatment, larvae were rinsed briefly and anesthetized with 0.016% tricaine in egg water supplemented with 0.2 mM PTU and mounted in 1% low-melting agarose for confocal imaging.
Image Acquisition, Processing, and Statistical Analysis
Zeiss LSM700 and Leica M205 FA were used for confocal microscopy and epifluorescence microscopy, respectively. ZEN 2009 software was used to assemble and analyze confocal images. Leica MZ16 stereomicroscope was used to obtain in situ images. Adobe Illustrator and Photoshop were used for assembling figures, schematics, and labels. GraphPad Prism 7 software was used to create and assemble quantitative data. Quantification for liver size, fluorescence, and cell counting was performed using the NIH ImageJ software. Unpaired two-tailed Student’s t-tests were used for most statistical analyses; for comparisons across >2 groups, one-way ANOVA was used. A value of p < 0.05 was considered statistically significant. Quantitative data are shown as means ± standard error of the mean (SEM).
RESULTS
Stat3 Inhibition Impairs LPC-Driven Liver Regeneration
We7 and others8,19 have previously characterized a zebrafish LPC-driven liver regeneration model where upon extensive hepatocyte ablation, BECs contribute to the repopulation of the liver. We utilized Tg(fabp10a:CFP-NTR) transgenic fish, which express the bacterial nitroreductase (NTR) enzyme fused with the cyan fluorescent protein (CFP) under the hepatocyte-specific, fabp10a, promoter7. Upon treatment of a nontoxic prodrug, Mtz, cells expressing NTR metabolize Mtz into a cytotoxic agent, inducing DNA interstrand cross-links and subsequent cell death specifically in NTR-expressing cells39. Since NTR-expressing hepatocytes undergo genetic-based apoptosis and can no longer contribute to the regenerative response, BECs contribute to liver regeneration in a multistep process. Initially, nearly all BECs dedifferentiate into HB-LCs, also termed LPCs, which strongly express Hnf4a (a hepatoblast/hepatocyte marker) and Prox1 (a hepatoblast/hepatocyte/BEC marker) in Alcam+ (a BEC marker) BEC-derived cells. Next, the LPCs proliferate and differentiate into either hepatocytes or BECs, concluding with the proliferation of the newly generated hepatocytes and BECs and the restoration of the lost liver mass. Using this model, we explored the implications of inflammatory signaling, specifically focusing on Stat3 and Socs3a, in LPC-driven liver regeneration.
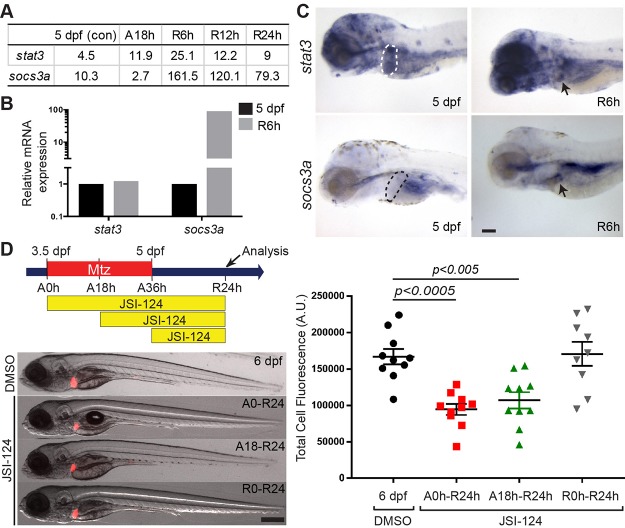
To induce hepatocyte ablation, fabp10a:CFP-NTR+ larvae were treated with Mtz from 3.5 to 5 days postfertilization (dpf). Following this 36-h treatment period (ablation; A36h), Mtz washout was considered as the initiation of regeneration (R0h). Based on our previously reported RNAseq analysis40, we found that stat3 and its negative feedback regulator, socs3a, were expressed in regenerating livers at R6h (Fig. 1A), which was confirmed by qPCR (Fig. 1B) and in situ hybridization (Fig. 1C, arrows). Since Socs3 is a direct target gene of Stat341, upregulation of socs3a in the regenerating livers (Fig. 1B) suggests active Stat3 signaling during the regeneration.
Figure 1.
Signal transducer and activator of transcription 3 (Stat3) inhibition impairs liver progenitor cell (LPC)-driven liver regeneration. (A) RNAseq data showing the expression levels of stat3 and socs3a in control 5 days postfertilization (dpf), ablated A18h, and regenerating R6h, R12h, and R24h livers. Their expression levels are shown in numbers indicating FPKM standing for fragments per kilobase of exon per million fragments mapped. (B) qPCR quantification showing the relative mRNA expression levels of stat3 and socs3a between nonablated 5 dpf and metronidazole (Mtz)-ablated R6h livers. (C) Whole-mount in situ hybridization (WISH) reveals stat3 and socs3a expression in nonablated 5-dpf livers (dashed lines) and Mtz-treated R6h livers (arrows). Lateral view, anterior to the left. (D) Epifluorescence images showing fabp10a:DsRed expression (red) in regenerating livers treated with dimethyl sulfoxide (DMSO) or JSI-124. Quantification of fabp10a:DsRed expression per liver area is shown. Total cell fluorescence (A.U., arbitrary units) considers both area of the liver and the fluorescence intensity. Scale bars: 100 μm; error bars: ± standard error of the mean (SEM).
To elucidate the role of Stat3 in LPC-driven liver regeneration, we utilized a JAK-specific kinase inhibitor, JSI-124, that blocks Stat3 activation42. JSI-124 has previously been used in zebrafish to block Stat3 signaling43,44. We first assessed the effect of chemically inhibiting Stat3 activation on liver size. To visualize the liver size in our regeneration setting, we utilized the Tg(fabp10a:DsRed) line that expresses DsRed in hepatocytes. Stat3 was inhibited from either A0h (at the start of Mtz treatment) to R24h, A18h to R24h, or R0h to R24h, at which time the liver size was examined. JSI-124 treatment significantly reduced the liver size at R24h from both the A0h and A18h, but not R0h, treatment (Fig. 1D), suggesting a positive role of Stat3 signaling during an earlier phase of LPC-driven liver regeneration, prior to R0h.
Stat3 Inhibition Reduces Liver Size Due to an LPC Proliferation Defect in Regenerating Livers
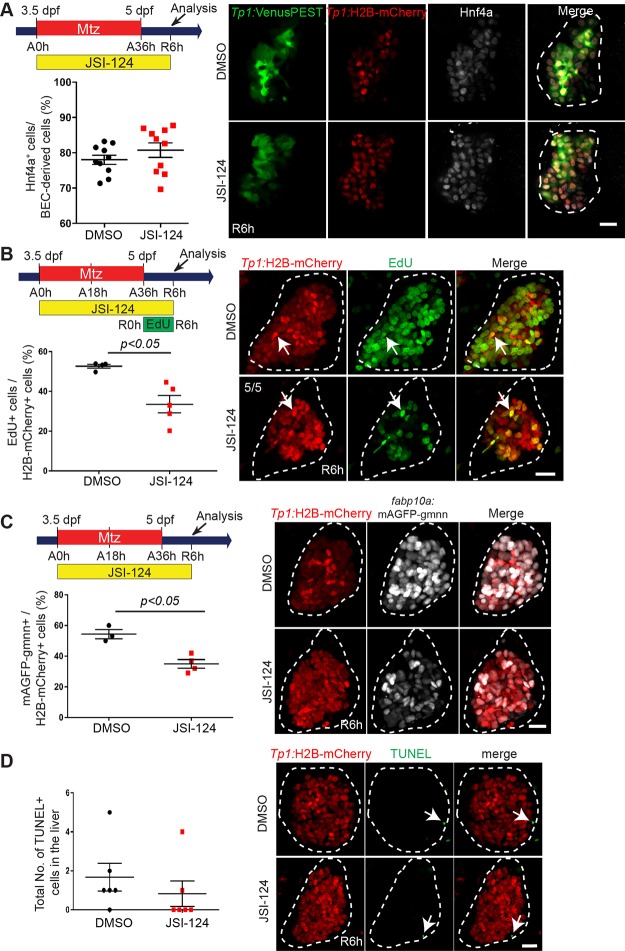
LPC-driven liver regeneration comprises three main steps: (1) BEC dedifferentiation into LPCs, (2) LPC proliferation, and (3) LPC-to-hepatocyte differentiation. First, we investigated the effect of Stat3 inhibition on BEC dedifferentiation. To mark BECs and BEC-derived cells, we used the Tg(Tp1:H2B-mCherry) line that expresses H2B-mCherry fusion proteins driven by the Tp1 promoter containing Notch response elements45. In the zebrafish liver, Notch activity is exclusively observed in BECs, and the prolonged stability of H2B-mCherry allows for marking BEC-derived cells even after Notch signaling is turned off. To inspect BEC dedifferentiation into LPCs, we examined the induction of Hnf4a in BECs or BEC-derived cells at R6h, indicative of BEC dedifferentiation. Typically, in the LPC-driven liver regeneration model, Hnf4a induction in BECs begins around A30h–A33h40, and by R6h, the Tp1:H2B-mCherry+ cells are referred to as LPCs or BEC-derived cells. Interestingly, in A0h-R6h JSI-124-treated larvae, the induction of Hnf4a in Tp1:H2B-mCherry+ cells (BEC-derived cells) was comparable to DMSO-treated control regenerating livers (Fig. 2A). These data indicate that BEC dedifferentiation in regenerating livers is unaffected by Stat3 inhibition.
Figure 2.
Stat3 inhibition reduces liver size due to an LPC proliferation defect in regenerating livers. (A) Confocal projection images showing the expression of Tp1:VenusPEST (green), Tp1:H2B-mCherry (red), and Hnf4a (gray) in regenerating livers at R6h. Quantification of the percentage of Hnf4a+ cells among biliary epithelial cell (BEC)-derived cells (H2B-mCherry+ cells) in regenerating livers is shown. (B) Confocal projection images showing Tp1:H2B-mCherry (red) expression and EdU (green) staining in regenerating livers at R6h. Quantification of the percentage of EdU+ cells among Tp1:H2B-mCherry+ cells (arrows) in the liver is shown. Numbers in the top left corner indicate the proportion of livers exhibiting the representative phenotype at R6h. (C) Confocal projection images showing Tp1:H2B-mCherry (red) and fabp10a:mAGFP-gmnn (gray) expression in regenerating livers at R6h. Quantification of the percentage of mAGFP-gmnn+ cells among Tp1:H2B-mCherry+ cells is shown. (D) Confocal projection images showing Tp1:H2B-mCherry (red) expression and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL; green) staining (arrows) in regenerating livers at R6h. Quantification of the total number of TUNEL+ cells per liver at R6h is shown. Dashed lines outline regenerating livers. Scale bars: 20 μm; error bars: ± SEM.
Next, we investigated whether the smaller liver size was due to a defect in LPC proliferation or cell death. To assess the first, we used the 5-ethynyl-2′-deoxyuridine (EdU) labeling and the cell cycle reporter line Tg(fabp10a:mAGFP-gmnn) 33. Based on the cell cycle oscillator phenomenon of Geminin proteins46, this transgenic line only labels hepatocytes in the S/G2/M phases of the cell cycle. In our zebrafish liver regeneration studies, this line labels proliferating LPCs at R6h33. Since we observed a gross phenotype at R24h (i.e., smaller liver size) following JSI-124 treatment from A0h to R24h, we hypothesized that any proliferation or cell death defect would occur prior to R24h. Following this logic, we conducted our EdU and transgenic cell cycle assays at R6h and R24h. Compared to control regenerating livers, JSI-124-treated regenerating livers displayed a significant decrease in LPC proliferation at R6h (Fig. 2B and C), but not at R24h (data not shown), indicative of a transient reduction in LPC proliferation.
Using the TUNEL assay, we assessed cell death in regenerating livers at R6h. Unlike the cell proliferation defect, there was no significant difference in TUNEL/Tp1:H2B-mCherry double-positive cells between control and JSI-124-treated regenerating livers (Fig. 2D). These data provide evidence that the smaller liver size is not due to an increase in cell death, but rather a defect in proliferation.
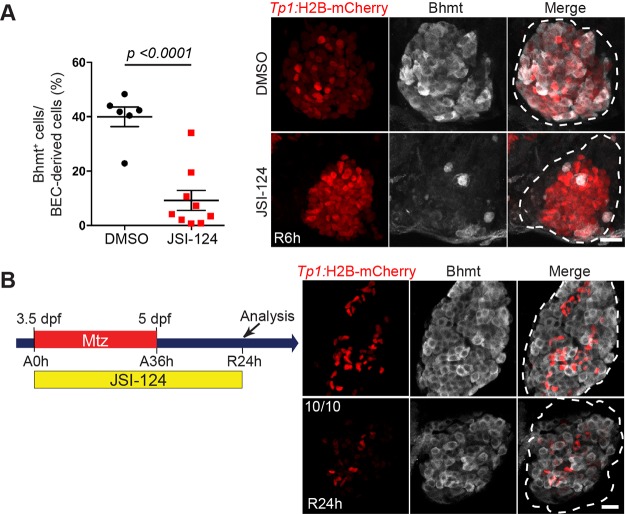
Stat3 Inhibition Temporarily Delays LPC-to-Hepatocyte Differentiation in Regenerating Livers
To examine the effect of Stat3 inhibition on LPC-to-hepatocyte differentiation, we used betaine-homocysteine S-methyltransferase (Bhmt) antibodies to mark newly generated hepatocytes40. At R6h, upon Stat3 inhibition, Bhmt expression was visibly reduced in 77% of the regenerating livers, indicative of an LPC-to-hepatocyte differentiation defect (Fig. 3A). Interestingly, this LPC-to-hepatocyte differentiation defect was temporary, as at a later stage, such as R24h, Bhmt expression in JSI-124-treated livers was comparable to that in control livers (Fig. 3B). These data indicate that Stat3 inhibition blocks LPC-to-hepatocyte differentiation; however, since Bhmt expression recovers by R24h, this blockage is transient. In addition to its role in mediating LPC proliferation, Stat3 also regulates the proper timing of LPC-to-hepatocyte differentiation.
Figure 3.
Stat3 inhibition temporarily delays LPC-to-hepatocyte differentiation in regenerating livers. (A) Confocal projection images showing Tp1:H2B-mCherry (red) and Bhmt (gray) expression in regenerating livers at R6h. Quantification of the percentage of Bhmt+ cells among BEC-derived cells in the regenerating livers is shown. (B) Confocal projection images showing Tp1:H2B-mCherry (red) and Bhmt (gray) expression in regenerating livers at R24h. The number of JSI-124-treated larvae exhibiting the representative phenotype shown at R24h is shown in the upper left corner. Dashed lines outline regenerating livers. Scale bars: 20 μm; error bars: ± SEM.
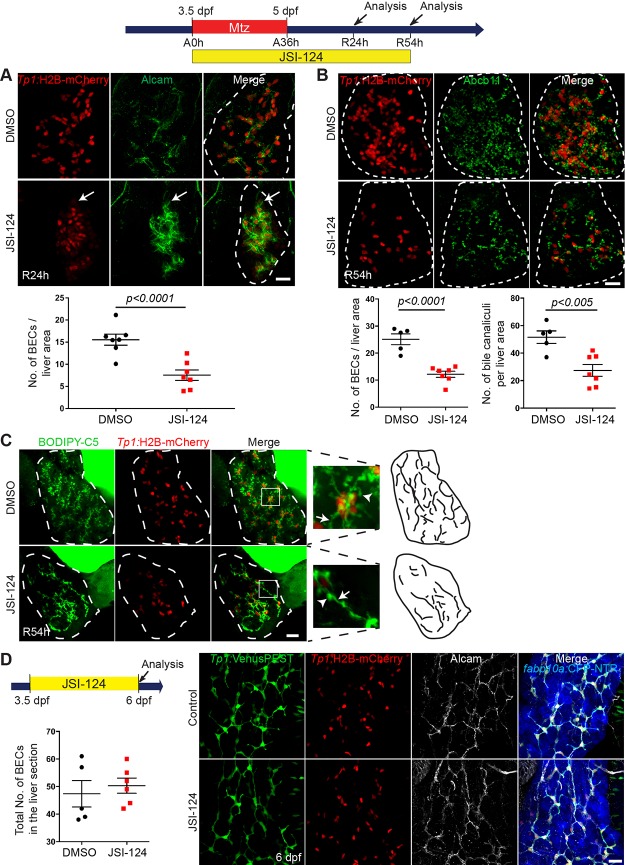
Stat3-Inhibited Regenerating Livers Contain Reduced BEC Numbers and Display Aberrant Intrahepatic Biliary Structure
In addition to the delayed differentiation of LPCs into hepatocytes in Stat3-inhibited livers, we observed a lower number of BECs in JSI-124-treated regenerating livers than in control regenerating livers, as assessed by Tp1:H2B-mCherry expression (Fig. 4A). In addition to the H2B-mCherry expression, we used another BEC marker, Alcam47, to clearly reveal BECs at R24h (Fig. 4A). Unlike the hepatocyte differentiation defect, which was temporary, the BEC number per liver area in JSI-124-treated regenerating livers remained low even later, such as R54h (Fig. 4B).
Figure 4.
Stat3-inhibited livers maintain low BEC numbers and display aberrant intrahepatic biliary structure. (A) Confocal projection images showing Tp1:H2B-mCherry (red) and Alcam (green) expression in regenerating livers at R24h. Quantification of the number of BECs (Alcam/H2B-mCherry double-positive cells) per liver is shown. For the count of BECs, the total number of BECs in the whole liver is divided by the liver area. Arrows point to weak H2B-mCherry+/Alcam+ cells, which were more prevalent in JSI-124-treated livers compared to controls. (B) Confocal projection images of regenerating livers at R54h showing Tp1:H2B-mCherry (red) and Abcb11 (green) expression. Quantification of the number of BECs (H2B-mCherry+ cells) and bile canaliculi per liver area is shown. (C) Confocal projection images showing BODIPY C5 staining (green) and Tp1:H2B-mCherry expression (red) at R54h. Inset boxes show a magnified image of the selected area. Arrows point to bile ductules; arrowheads point to bile canaliculi. (D) Confocal projection images showing the expression of Tp1:H2B-mCherry (red), Tp1:VenusPEST (green), fabp10a:CFP-NTR (blue), and Alcam (gray) in nonablated livers treated with DMSO or JSI-124. Quantification of the number of BECs (Alcam+/H2B-mCherry+ cells) per liver is shown. Dashed lines outline regenerating livers. Scale bars: 20 μm; error bars: ± SEM.
We further examined the impact of delayed LPC-to-hepatocyte differentiation and reduced BEC number by investigating hepatocyte polarity and biliary morphogenesis. Typically expressed on the apical side of the hepatocyte membrane, the bile salt export pump marker, ATP-binding cassette subfamily B member 11 (Abcb11), marks bile canaliculi in hepatocytes48,49. The number of Abcb11+ cells per liver area was significantly decreased in JSI-124-treated livers compared to DMSO-treated controls (Fig. 4B). We confirmed the Abcb11 phenotype with the BODIPY C5 lipid analog assay, which allows for the visualization of the intrahepatic biliary network as the processed lipids pass through the biliary conduits38. Compared to the control regenerating livers, JSI-124-treated regenerating livers had shorter and round-shaped bile canaliculi (Fig. 4C, arrowheads) and reduced density of biliary conduits (Fig. 4C, cartoons). JSI-124 treatment in nonablated control livers, however, had no effect on liver morphology or BEC number (Fig. 4D), suggesting the regeneration-restricted role of Stat3 in the zebrafish liver.
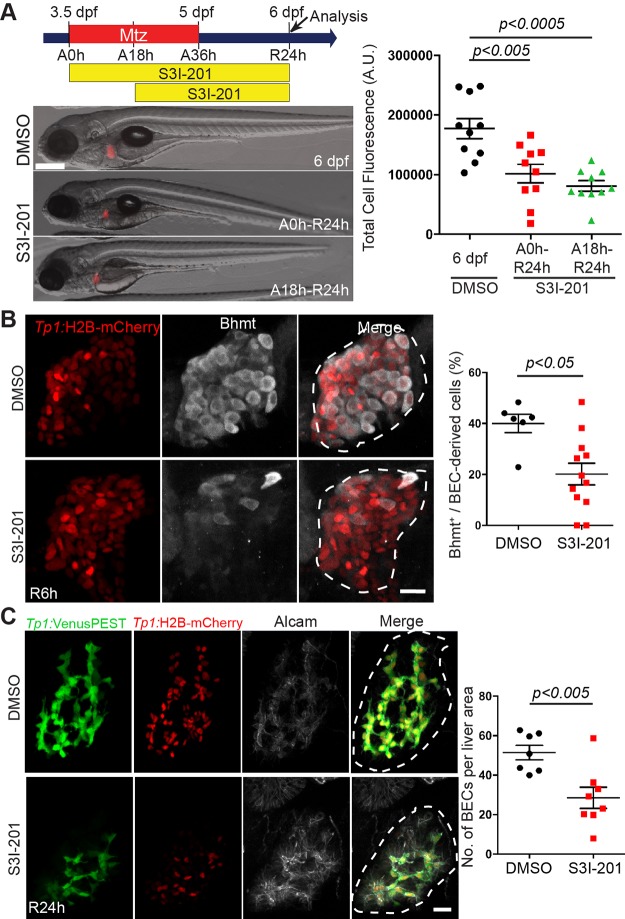
We further validated the effect of blocking Stat3 using a different inhibitor, S3I-201, which blocks Stat3 dimerization and subsequent Stat3 activation50. Similar to the JSI-124 liver phenotypes, S3I-201-treated larvae also exhibited a significantly smaller regenerating liver size at R24h (Fig. 5A), a delay in LPC-to-hepatocyte differentiation at R6h (Fig. 5B), and a reduced BEC number at R24h (Fig. 5C). Altogether, these data implicate Stat3 as an important regulator in the timing for LPC-to-hepatocyte differentiation and in the establishment of a proper BEC population during LPC-driven liver regeneration.
Figure 5.
Stat3 inhibition by S3I-201 impairs LPC-driven liver regeneration. (A) Epifluorescence images showing fabp10a:DsRed expression (red) in the regenerating livers treated with DMSO or S3I-201 at the indicated time points. Quantification of fabp10a:DsRed expression per liver area is shown. Total cell fluorescence considers both area of the liver and the fluorescence intensity. (B) Confocal projection images showing Tp1:H2B-mCherry (red) and Bhmt (gray) expression in regenerating livers at R6h. Quantification of the percentage of Bhmt+ cells among BEC-derived cells is shown. (C) Confocal projection images showing the expression of Tp1:VenusPEST (green), Tp1:H2B-mCherry (red), and Alcam (gray) in regenerating livers at R24h. Quantification of the number of BECs (H2B-mCherry+ cells) per liver area is shown. Dashed lines outline regenerating livers. Scale bars: 100 μm (A), 20 μm (B, C); error bars: ± SEM.
A Subset of stat3 Homozygous Mutants Exhibit Fewer BECs and a Delay in LPC-to-Hepatocyte Differentiation During LPC-Driven Liver Regeneration
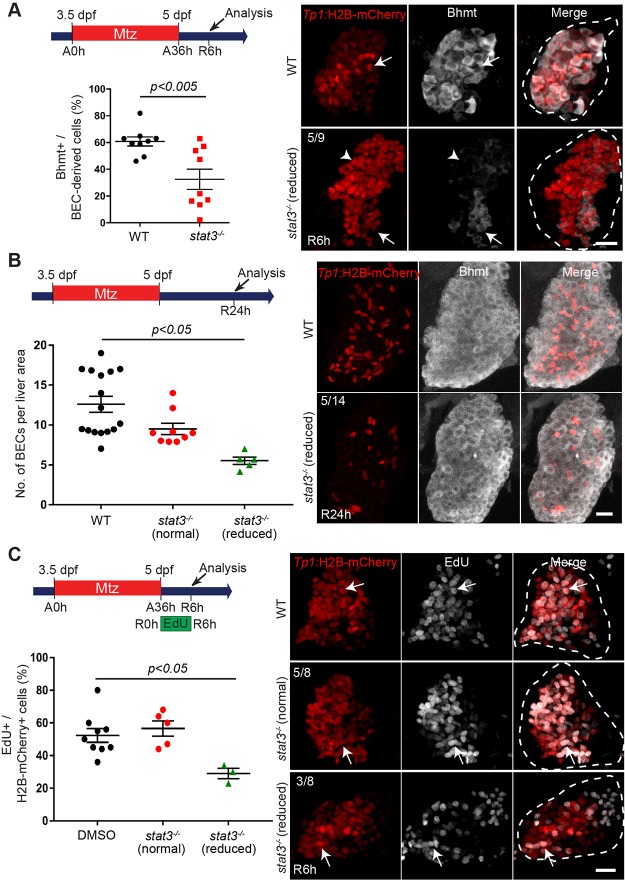
To determine whether stat3 mutants phenocopied the aberrant liver regeneration phenotypes observed in JSI-124-treated larvae, we examined LPC-driven liver regeneration in stat3 −/− mutants29, focusing on three main phenotypes: (1) LPC-to-hepatocyte differentiation, (2) BEC number, and (3) LPC proliferation. First, similar to the JSI-124-treated larvae, stat3 −/− mutants exhibited reduced Bhmt expression at R6h (Fig. 6A), indicative of an initial defect in LPC-to-hepatocyte differentiation. However, only ∼55% of stat3 −/− mutants (n = 9) exhibited decreased Bhmt expression. The other 45% of stat3 −/− mutants showed no significant difference in Bhmt expression compared to wild-type larvae. Moreover, as in the JSI-124-treated livers, hepatocyte differentiation is not completely blocked but rather delayed as Bhmt expression at R24h was unaffected between wild-type and stat3 −/− mutants (Fig. 6B).
Figure 6.
A subset of stat3 homozygous mutants exhibit reduced number of BECs and a delay in LPC-to-hepatocyte differentiation during LPC-driven liver regeneration. (A) Confocal projection images showing Tp1:H2B-mCherry (red) and Bhmt (gray) expression in regenerating livers of wild-type or stat3 homozygous mutants at R6h. Quantification of the percentage of Bhmt+ cells among H2B-mCherry+ cells is shown. The proportion of regenerating livers exhibiting the Bhmt phenotype in stat3 mutants is shown in the top left corner. Arrows point to Bhmt+ and H2B-mCherry+ cells. Arrowheads point to Bhmt- and H2B-mCherry+ cells. (B) Confocal projection images showing Tp1:H2B-mCherry (red) and Bhmt (gray) expression at R24h in wild-type and stat3 mutants. Quantification of the number of strong H2B-mCherry+ cells per liver area is shown. The proportion of stat3 −/− mutants exhibiting the reduced BEC number phenotype is shown in the upper left corner. (C) Confocal projection images showing Tp1:H2B-mCherry (red) expression and EdU (gray) staining in regenerating R6h livers of wild-type and stat3 −/− mutants. Numbers in the top left corner indicate the proportion of stat3 mutant livers exhibiting reduced proliferation (n = 3/8) versus nonaffected stat3 mutants (n = 5/8) compared to wild-type larvae. Quantification of the percentage of EdU+ cells among H2B-mCherry+ cells in the regenerating liver is shown. Arrows point to EdU/H2B-mCherry double-positive cells; dashed lines outline regenerating livers. Scale bars: 20 μm; error bars: ± SEM.
At R24h, ∼35% of the stat3 −/− mutants (n = 14) exhibited a low BEC number in the regenerating livers (Fig. 6B). Moreover, a significant decrease in the percentage of EdU/Tp1:H2B-mCherry double-positive cells was observed at R6h in a subset (37%; n = 8) of stat3 −/− mutants, indicating an LPC proliferation defect (Fig. 6C). Altogether, data from stat3 mutant analyses suggest that stat3 partially regulates LPC proliferation and LPC-to-hepatocyte differentiation during LPC-driven liver regeneration.
socs3a Homozygous Mutants Exhibit a Mild but Significant Defect in BEC Number During LPC-Driven Liver Regeneration
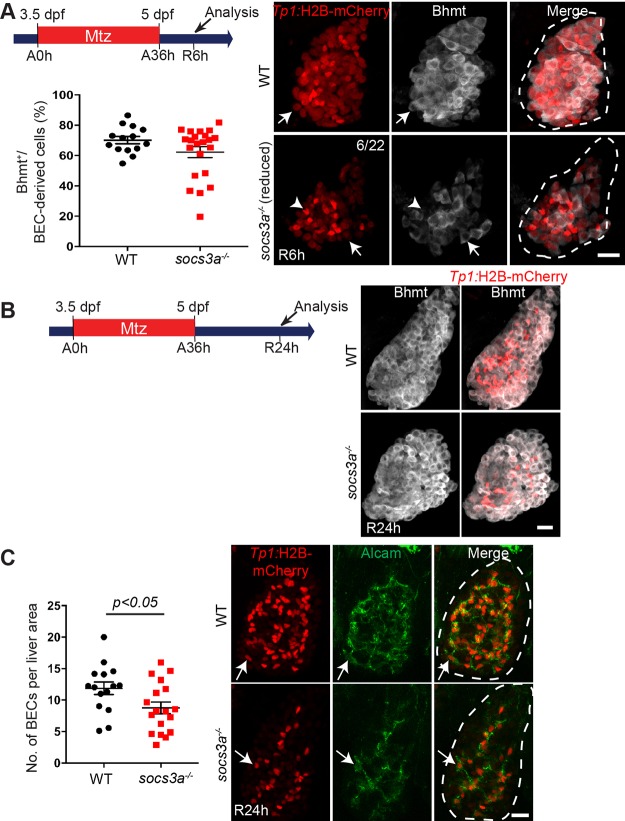
Since Socs3a acts as a downstream negative regulator of the Stat3 signaling pathway51, we speculated that absence of socs3a would allow us to study the effects of Stat3 hyperactivation on LPC-driven liver regeneration. Using TALEN genome editing technology, we generated socs3a mutants with a 61-base deletion. The socs3a mutant larvae exhibited no liver developmental defects and grew normally to adults. Interestingly, as assessed by Bhmt expression, LPC-to-hepatocyte differentiation was affected in ∼30% of socs3a −/− mutants (n = 22) at R6h (Fig. 7A), but not affected at all at R24h (Fig. 7B). In addition, the mutant regenerating livers had a significant decrease in BEC number compared to wild type at R24h (Fig. 7C). Altogether, our data from socs3a mutant analyses suggest that similar to Stat3, Socs3a also partially controls LPC-to-hepatocyte differentiation and establishment of a proper BEC population during LPC-driven liver regeneration.
Figure 7.
socs3a homozygous mutants exhibit a mild but significant defect in BEC number during LPC-driven liver regeneration. (A) Confocal projection images showing Tp1:H2B-mCherry (red) and Bhmt (gray) expression in the regenerating livers of wild-type and socs3a mutants at R6h. Numbers in the top right corner indicate the proportion of socs3a mutants exhibiting a reduced number of BECs at R6h (n = 6/22). Quantification of the percentage of Bhmt+ cells among H2B-mCherry+ cells is shown. Arrows point to Bhmt+ and H2B-mCherry+ cells. Arrowheads point to Bhmt- and H2B-mCherry+ cells. (B) Confocal projection images of wild-type and socs3a mutant regenerating livers exhibiting Tp1:H2B-mCherry (red) and Bhmt (gray) expression at R24h. (C) Confocal projection images showing Tp1:H2B-mCherry (red) and Alcam (green) expression in the regenerating livers of wild-type and socs3a mutants at R24h. Quantification of the number of BECs (H2B-mCherry/Alcam double-positive cells) per liver area is shown. Arrows point to H2B-mCherry/Alcam double-positive cells. Dashed lines outline regenerating livers. Scale bars: 20 μm; error bars: ± SEM.
DISCUSSION
The signaling pathways regulating LPC proliferation and differentiation during LPC-driven liver regeneration remain poorly understood. In this study, we sought to determine the role of Stat3 in LPC-driven liver regeneration by modulating Stat3 signaling using chemical inhibitors, JSI-124 and S3I-201, as well as stat3 and socs3a mutants. Chemically inhibiting Stat3 significantly reduced the size of regenerating livers at R24h. To elucidate the process of LPC-driven liver regeneration affected by Stat3 inhibition, we examined (1) BEC dedifferentiation, (2) LPC proliferation, and (3) LPC differentiation into hepatocytes. First, we found that BEC dedifferentiation during liver regeneration was unaffected in JSI-124-treated livers. We further noted that Stat3 inhibition decreased the number of BECs in regenerating livers at R24h, and this decrease was still maintained later at R54h. Additionally, our data revealed that during liver regeneration, in Stat3 inhibitor-treated animals, livers exhibited a decrease in LPC proliferation at R6h, but not at R24h, which helps to explain the decreased liver size at R24h and recovery of the liver size at later time points. Last, Stat3 inhibition resulted in a delay in LPC-to-hepatocyte differentiation during LPC-driven liver regeneration.
Given no current marker that can clearly distinguish between LPCs and BECs, we utilized the induction of fabp10a:rasGFP in BECs to show that nearly all BECs dedifferentiate into LPCs in the zebrafish hepatocyte ablation model40. Since the induction of Hnf4a in BECs/BEC-derived cells, which was used as a marker for BEC dedifferentiation, was found to be comparable between DMSO- and JSI-124-treated larvae, we concluded that BEC dedifferentiation is not affected upon Stat3 loss. Thus, since BEC dedifferentiation is occurring as expected, we can speculate that the reduced BEC number in the JSI-124-treated larvae at R24h is not due to a BEC dedifferentiation defect. Instead, it is more likely due to both the reduced LPC-to-BEC differentiation and reduced LPC proliferation. This reduction in BEC number may also affect the proper formation of intrahepatic bile ducts later, such as at R54h.
In our zebrafish LPC-driven liver regeneration model, stat3 was expressed in regenerating livers during the early phases of liver regeneration (R6h), when the regenerating liver predominantly consists of LPCs. In the 2-AAF/PHx rat model of oval cell activation, in which 2-acetylaminfluorene (2-AAF) is administered prior to PHx to block hepatocyte proliferation, Stat3 was highly upregulated in the oval cell population that correlated with the highest proliferative potential25. Depending on the cell type and injury stimulus, Stat3 plays a critical role in several biological processes, including proliferation, apoptosis, and inflammation. Previous studies have implicated Stat3 in the regulation of myogenic differentiation in skeletal muscles. During muscle repair, Stat3 activation in satellite cells drives these muscle stem cells to choose differentiation over self-renewal52,53, which partly supports a delayed LPC-to-hepatocyte differentiation in Stat3-inhibited regenerating livers. In a separate study of two mouse models, a muscle satellite cell (MuSC)-specific Stat3 knockout and Stat3/dystrophin double knockout, cell proliferation of the MuSCs was decreased54. This supports our finding that inhibition of Stat3 resulted in a proliferation defect in LPCs at R6h during LPC-driven liver regeneration. Therefore, we can speculate that the smaller liver size at R24h is a result of the lower number of LPCs established at R6h, resulting in a subsequent lower number of LPCs available for hepatocyte differentiation.
Several mouse studies have also examined the role of Stat3 in liver regeneration. For example, following PHx in liver-specific Stat3 knockout mice, the mortality rate was significantly increased during liver regeneration24. This increase was speculated to be due to a defect in the Stat3-dependent acute phase response; this response is activated following liver injury and involves the induction of survival-related proteins55,56. In a separate model of liver injury (i.e., CCl4), although liver-specific Stat3 knockout mice had a slight decrease in hepatocyte proliferation, they displayed no difference in survival or necrosis compared to wild-type littermates24.
In our LPC-driven liver regeneration model, we discovered a reduction in Bhmt expression in a majority of JSI-124-treated regenerating livers at R6h, which indicates an LPC-to-hepatocyte differentiation defect. However, at R24h, no difference was observed in Bhmt expression between control and JSI-124-treated regenerating livers, indicating a delay in hepatocyte differentiation rather than a complete blockage. Possibly, Stat3 is one of several important regulators implicated in hepatocyte differentiation; hence, upon Stat3 inhibition, other compensatory mechanisms may be activated to counteract the Stat3 deficiency. Such a phenomenon was detected in liver-specific Stat3 knockout mice: STAT1, which is minimally expressed in noninjured livers, had increased expression levels following PHx57. Moreover, we observed that both stat3 knockout and Stat3 hyperactivation (socs3a knockout) conditions in zebrafish displayed similar phenotypes in LPC-driven liver regeneration, including delayed LPC differentiation. We propose that perturbations that disturb the optimal level of Stat3 activity, either through reduction (stat3 mutant) or through enhancement (socs3a mutant), can lead to similar defects. Such a phenomenon, where loss of function and overexpression of a particular gene exhibit identical phenotypes, has been observed in multiple proteins, including SPT558. In addition, perturbation, via knockout or overexpression, of the kinase SRPK1, a downstream target of AKT, was found to have the same tumorigenic phenotype in mouse embryonic fibroblasts59.
Moreover, data from our stat3 and socs3a mutant analyses suggest that stat3 and socs3a only partially recapitulate the phenotypes observed in Stat3 chemically inhibited livers. One possibility is that since complete loss of a gene’s function can prove harmful for the organism, cells may offset a permanent, genetic loss of a gene with compensatory mechanisms, which are not employed upon temporary chemical inhibition. A similar occurrence of incomplete phenotypic reproducibility between temporary versus genetic/permanent inhibition of a gene’s function is seen in morpholino versus genetic knockouts60. Second, although JSI-124 blocks Stat3 signaling, it does so by targeting the upstream activator of Stat3, namely, JAK kinases. In contrast, in the stat3 mutants, stat3 expression is lost by direct, targeted mutation at the stat3 locus. Because of the temporary, acute, and indirect nature of Stat3 inhibition in JSI-124-treated larvae, compared to the permanent, genetic, and direct loss of Stat3 in mutants, the chemically treated cells may not have employed adequate counterbalancing measures to compensate for the loss of an important signaling pathway (i.e., Stat3).
Although we did not address the upstream activators or downstream mediators of the Stat3 signaling pathway, future studies can focus on identifying these factors and their role in LPC proliferation and differentiation. Factors such as the IL cytokines may be important activators of the Stat3/Socs3 pathway during LPC-driven liver regeneration. In particular, in the CDE diet-fed mouse model of liver injury and oval cell activation, IL-6 levels were upregulated, and IL-6 knockout mice displayed a reduction in total oval cell numbers21. Moreover, when CDE diet-fed mice were stimulated with IL-6, Socs3 expression was increased in the LPCs21, raising a possibility that IL-6 or its homolog may be the upstream activator of Stat3 signaling during LPC-driven liver regeneration.
ACKNOWLEDGMENTS
We thank Jinrong Peng for the anti-Bhmt antibody, Ken Poss for the stat3 in situ probe, and Neil Hukriede and Michael Tsang for the discussions. This work was supported by grants from the NIH to D.S. (DK101426), M.K. (F31DK105714), and L.S.K. (R35GM118179), and from the Chinese Academy of Sciences and National Natural Science Foundation of China to Y.S. (31671501). This work was funded by NIH grants to D.S. (R01DK101426), L.S.K. (R35GM118179), and M.K. (F31DK105714) and by the Chinese Academy of Sciences and National Natural Science Foundation of China to Y.S. (31671501).
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997;276(5309):60–6. [DOI] [PubMed] [Google Scholar]
- 2. Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 2014;14(5):561–74. [DOI] [PubMed] [Google Scholar]
- 4. Okabe M, Tsukahara Y, Tanaka M, Suzuki K, Saito S, Kamiya Y, Tsujimura T, Nakamura K, Miyajima A. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 2009;136(11):1951–60. [DOI] [PubMed] [Google Scholar]
- 5. Paku S, Schnur J, Nagy P, Thorgeirsson SS. Origin and structural evolution of the early proliferating oval cells in rat liver. Am J Pathol. 2001;158(4):1313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, and others. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 2015;17(8):971–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 2014;146(3):776–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 2014;146(3):789–800 e8. [DOI] [PubMed] [Google Scholar]
- 9. Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014;15(5):605–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stueck AE, Wanless IR. Hepatocyte buds derived from progenitor cells repopulate regions of parenchymal extinction in human cirrhosis. Hepatology 2015;61(5):1696–707. [DOI] [PubMed] [Google Scholar]
- 11. Kochanek KD, Murphy SL, Xu JQ, B T-V. Deaths: Final data for 2014. National Vital Statistics Reports: National Center for Health Statistics; 2016. [PubMed] [Google Scholar]
- 12. Organ Procurement and Transplantation Network (OPTN) and Scientific Registry of Transplant Recipients (SRTR). OPTN/SRTR 2012 Annual Data Report. Rockville, MD: Department of Health and Human Services, Health Resources and Services Administration; 2014. [Google Scholar]
- 13. Weng HL, Cai X, Yuan X, Liebe R, Dooley S, Li H, Wang TL. Two sides of one coin: Massive hepatic necrosis and progenitor cell-mediated regeneration in acute liver failure. Front Physiol. 2015;6:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sancho-Bru P, Altamirano J, Rodrigo-Torres D, Coll M, Millán C, José Lozano J, Miquel R, Arroyo V, Caballería J, Ginès P, and others. Liver progenitor cell markers correlate with liver damage and predict short-term mortality in patients with alcoholic hepatitis. Hepatology 2012;55(6):1931–41. [DOI] [PubMed] [Google Scholar]
- 15. Nobili V, Carpino G, Alisi A, Franchitto A, Alpini G, De Vito R, Onori P, Alvaro D, Gaudio E. Hepatic progenitor cells activation, fibrosis, and adipokines production in pediatric nonalcoholic fatty liver disease. Hepatology 2012;56(6):2142–53. [DOI] [PubMed] [Google Scholar]
- 16. Clouston AD, Powell EE, Walsh MJ, Richardson MM, Demetris AJ, Jonsson JR. Fibrosis correlates with a ductular reaction in hepatitis C: Roles of impaired replication, progenitor cells and steatosis. Hepatology 2005;41(4):809–18. [DOI] [PubMed] [Google Scholar]
- 17. Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol. 1999;154(2):537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang W, Wang C, Lin Y, Liu Q, Yu LX, Tang L, Yan HX, Fu J, Chen Y, Zhang HL, and others. OV6+ tumor-initiating cells contribute to tumor progression and invasion in human hepatocellular carcinoma. J Hepatol. 2012;57(3):613–20. [DOI] [PubMed] [Google Scholar]
- 19. Huang M, Chang A, Choi M, Zhou D, Anania FA, Shin CH. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology 2014;60(5):1753–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O’Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, and others. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 2017;547(7663):350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeoh GC, Ernst M, Rose-John S, Akhurst B, Payne C, Long S, Alexander W, Croker B, Grail D, Matthews VB. Opposing roles of gp130-mediated STAT-3 and ERK-1/ 2 signaling in liver progenitor cell migration and proliferation. Hepatology 2007;45(2):486–94. [DOI] [PubMed] [Google Scholar]
- 22. Hemmann U, Gerhartz C, Heesel B, Sasse J, Kurapkat G, Grötzinger J, Wollmer A, Zhong Z, Darnell JE, Graeve L, and others. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. II. Src homology SH2 domains define the specificity of stat factor activation. J Biol Chem. 1996;271(22):12999–3007. [DOI] [PubMed] [Google Scholar]
- 23. Zhong Z, Wen Z, Darnell JE. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994;264(5155):95–8. [DOI] [PubMed] [Google Scholar]
- 24. Moh A, Iwamoto Y, Chai GX, Zhang SS, Kano A, Yang DD, Zhang W, Wang J, Jacoby JJ, Gao B, and others. Role of STAT3 in liver regeneration: Survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest. 2007;87(10):1018–28. [DOI] [PubMed] [Google Scholar]
- 25. Sánchez A, Factor VM, Schroeder IS, Nagy P, Thorgeirsson SS. Activation of NF-kappaB and STAT3 in rat oval cells during 2-acetylaminofluorene/partial hepatectomy-induced liver regeneration. Hepatology 2004;39(2):376–85. [DOI] [PubMed] [Google Scholar]
- 26. Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 1999;4(6):339–51. [DOI] [PubMed] [Google Scholar]
- 27. Riehle KJ, Campbell JS, McMahan RS, Johnson MM, Beyer RP, Bammler TK, Fausto N. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med. 2008;205(1):91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene: University of Oregon Press; 2000. [Google Scholar]
- 29. Liu Y, Sepich DS, Solnica-Krezel L. Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 2017;13(2):e1006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korzh S, Pan X, Garcia-Lecea M, Winata CL, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 2012;139(9):1557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39(3):397–402. [DOI] [PubMed] [Google Scholar]
- 33. Ko S, Choi TY, Russell JO, So J, Monga SPS, Shin D. Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration. J Hepatol. 2016;64(2):316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29(8):699–700. [DOI] [PubMed] [Google Scholar]
- 35. Choi TY, Khaliq M, Ko S, So J, Shin D. Hepatocyte-specific ablation in zebrafish to study biliary-driven liver regeneration. J Vis Exp. 2015(99):e52785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22(3):288–99. [DOI] [PubMed] [Google Scholar]
- 37. de Groh ED, Swanhart LM, Cosentino CC, Jackson RL, Dai W, Kitchens CA, Day BW, Smithgall TE, Hukriede NA. Inhibition of histone deacetylase expands the renal progenitor cell population. J Am Soc Nephrol. 2010;21(5):794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev Biol. 2011;360(2):276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: A spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3(6):948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi TY, Khaliq M, Tsurusaki S, Ninov N, Stainier DYR, Tanaka M, Shin D. Bone morphogenetic protein signaling governs biliary-driven liver regeneration in zebrafish through tbx2b and id2a. Hepatology 2017;66(5):1616–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: Characterization of the murine SOCS-3 promoter. Proc Natl Acad Sci USA 1999;96(12):6964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63(6):1270–9. [PubMed] [Google Scholar]
- 43. Elsaeidi F, Bemben MA, Zhao XF, Goldman D. Jak/Stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of Socs3 and Sfpq. J Neurosci. 2014;34(7):2632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao XF, Wan J, Powell C, Ramachandran R, Myers MG, Goldman D. Leptin and IL-6 family cytokines synergize to stimulate Müller glia reprogramming and retina regeneration. Cell Rep. 2014;9(1):272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8(6):e1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, and others. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008;132(3):487–98. [DOI] [PubMed] [Google Scholar]
- 47. Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18(20):1565–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239(3):855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, Meier PJ. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273(16):10046–50. [DOI] [PubMed] [Google Scholar]
- 50. Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, and others. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA 2007;104(18):7391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Duhé RJ, Wang LH, Farrar WL. Negative regulation of Janus kinases. Cell Biochem Biophys. 2001;34(1):17–59. [DOI] [PubMed] [Google Scholar]
- 52. Sun L, Ma K, Wang H, Xiao F, Gao Y, Zhang W, Wang K, Gao X, Ip N, Wu Z. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J Cell Biol. 2007;179(1):129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang K, Wang C, Xiao F, Wang H, Wu Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem. 2008;283(49):34029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu H, Xiao F, Wang G, Wei X, Jiang L, Chen Y, Zhu L, Wang H, Diao Y, Ip NY, and others. STAT3 regulates self-renewal of adult muscle satellite cells during injury-induced muscle regeneration. Cell Rep. 2016;16(8):2102–15. [DOI] [PubMed] [Google Scholar]
- 55. Akira S. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 2000;19(21):2607–11. [DOI] [PubMed] [Google Scholar]
- 56. Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol Cell Biol. 2001;21(5):1621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li W, Liang X, Kellendonk C, Poli V, Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem. 2002;277(32):28411–7. [DOI] [PubMed] [Google Scholar]
- 58. Clark-Adams CD, Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7(2):679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang P, Zhou Z, Hu A, Ponte de Albuquerque C, Zhou Y, Hong L, Sierecki E, Ajiro M, Kruhlak M, Harris C, and others. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol Cell 2014;54(3):378–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015;524(7564):230–3. [DOI] [PubMed] [Google Scholar]