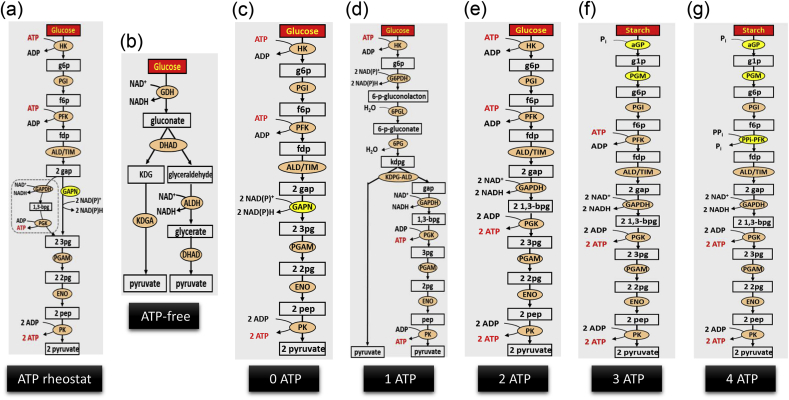
Fig. 1.
Pathway design for ATP regeneration or balancing. The enzymes are GDH, glucose dehydrogenase; DHAD, dihydroxy acid dehydratase; KDGA, 2-keto-3-desoxygluconate aldolase; ALDH glyceraldehyde dehydrogenase; HK, hexokinase; PGI, phosphoglucose isomerase; PFK, 6-phosphofructokinase; ALD, fructosebisphosphate aldolase; TIM, triosephosphate isomerase; GAPN, non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase; PGAM, cofactor-independent phosphoglycerate mutase; ENO, enolase; PK, pyruvate kinase; G6PDH, glucose 6-phosphate dehydrogenase; 6PGL, 6-phosphogluconolactonase; 6PG, 6-phosphogluconate dehydratase; KDPG-ALD, 2-keto-3-deoxy-phosphogluconate aldolase; GAPDH glyceraldehyde 3-phosphate dehydrogenase; PGK phosphoglycerate dehydrogenase; aGP, alpha-glucan phosphorylase; PGM, phosphoglucomutase; PPi-PFK, pyrophosphate-dependent fructose-6-phosphate 1-phosphotransferase. Metabolites are KDG, 2-keto-3-deoxygluconate; g6p, glucose 6-phosphate; f6p, fructose 6-phosphate; fdp, fructose 1,6-diphosphate; gap, glyceraldehyde 3-phosphate; 3pg, 3-phosphoglycerate; 2pg, 2-phosphoglycerate; pep, phosphoenolpyruvate; kdpg, 2-keto-3-deoxy-6-phosphogluconate; 1,3-bpg, 1,3-diphosphoglycerate; dhap, dihydroxacetone phosphate.