Abstract
Macrophages are well characterized as immune cells. However, in recent years, a multitude of non-immune functions have emerged many of which play essential roles in a variety of developmental processes (Wynn, Chawla and J. W. Pollard, 2013; DeFalco et al., 2014). In adult animals, macrophages are derived from circulating monocytes originating in the bone marrow, but much of the tissue-resident population arise from erythro-myeloid progenitors (EMPs) in the extra-embryonic yolk sac, appearing around the same time as primitive erythroblasts (Schulz et al., 2012; Kierdorf et al., 2013; McGrath, Frame and Palis, 2015; Gomez Perdiguero et al., 2015; Mass et al., 2016). Of particular interest to our group, macrophages have been shown to act as pro-angiogenic regulators during development (Owen and Mohamadzadeh, 2013; DeFalco et al., 2014; Hsu et al., 2015), but there is still much to learn about these early cells.
The goal of the present study was to isolate and expand progenitors of yolk-sacderived Embryonic Macrophages (EMs) in vitro to generate a new platform for mechanistic studies of EM differentiation. To accomplish this goal, we isolated pure (>98%) EGFP+ populations by flow cytometry from embryonic day 9.5 (E9.5) Csf1r-EGFP+/tg mice, then evaluated the angiogenic potential of EMs relative to Bone Marrow-Derived Macrophages (BMDMs). We found that EMs expressed more pro-angiogenic and less pro-inflammatory macrophage markers than BMDMs. EMs also promoted more endothelial cell (EC) cord formation in vitro, as compared to BMDMs in a manner that required direct cell-to-cell contact. Importantly, EMs preferentially matured into microglia when co-cultured with mouse Neural Stem/Progenitor Cells (NSPCs). In conclusion, we have established a protocol to isolate and propagate EMs in vitro, have further defined specialized properties of yolk-sac-derived macrophages, and have identified EM-EC and EM-NSPC interactions as key inducers of EC tube formation and microglial cell maturation, respectively.
Keywords: Yolk sac, Macrophages, Microglia, Angiogenesis, Endothelial Cells, Neural Stem/Progenitor Cells
INTRODUCTION
Beside their key role in immune response, macrophages have been shown to mediate the development and maintenance of normal structure and function of several tissues including the vascular system. For instance, macrophages were previously shown to regulate vascular complexity during retinal development by secreting VEGF-C, which activates VEGFR3 on vascular endothelial cells and reinforces Notch expression (Tammela et al., 2011). Additionally, previous studies have suggested that macrophages function as cellular chaperones to enhance vascular anastomosis in the developing hindbrain and retina by mediating the fusion of endothelial tip cells (Fantin et al., 2010; Rymo et al., 2011). Others have found that macrophages function in vascular remodeling and regression such as in the case of the transient hyaloid vasculature of the eye. Here, resident macrophages mediate vascular endothelial cell apoptosis via the cooperation between the angiopoietin-2 and Wnt7β pathways (Rao et al., 2007). In addition, recent work has indicated that macrophages promote pupillary membrane capillary regression by engulfing endothelial cell membrane particles emanating from the pupillary membrane vasculature (Poché et al., 2015). Together, these studies suggest that macrophages play various roles during the development and remodeling of the vascular system, but the exact mechanisms underlying this functional diversity are not fully understood.
Beyond development, macrophages have also been implicated in pathological angiogenesis of several diseases such as cancer. For instance, tumor associated macrophages (TAMs) play an important role in tumor angiogenesis as they regulate the angiogenic switch leading to increased tumor vascularization, which is required for the transition to the malignant state (Lin et al., 2006). Additionally, studies have shown that TAMs can secrete many pro-angiogenic factors and proteases. TAMs are also characterized by the expression of the mannose receptor 1 (Mrc1), which is also highly expressed on embryonic macrophages during development (Takahashi et al., 1998; Lin and J. W. Pollard, 2007; Mazzieri et al., 2011). These data are in line with the previous finding that TAMs and EMs share a common gene signature suggesting overlapping functions possibly at the level of angiogenic potential (Pucci et al., 2009). While these studies investigated the function of adult macrophages in a tumor environment, our project aimed at elucidating the functional properties of yolk sac-derived embryonic macrophages (EMs) isolated from the embryo proper.
The many described roles for macrophages point to tremendous functional heterogeneity, but also plasticity. Many groups have described the ability of macrophages to respond to altered cellular environments by dramatically changing their functional role. For example, in the broadest sense, the presence of specific signaling molecules are well known to instruct macrophages to adopt either the so-called M1 (pro-inflammatory) identity versus the M2 (anti-inflammatory) identity (Jetten et al., 2014). Importantly, EMs have been shown to express M2 markers such as Mrc1 and Arginase 1, suggesting they are more similar to the M2, versus the M1, classification scheme (Takahashi et al., 1998; Rőszer, 2015). Additionally, M2 macrophages have been reported to exhibit pro-angiogenic activities. However, it is important to recognize that within these two very general classifications of macrophages, other levels of functional variation likely exist along a continuum (Ovchinnikov, 2008; Pucci et al., 2009; Mazzieri et al., 2011; Jetten et al., 2014).
In addition to environmental cues, macrophages may also be influenced by developmental origin and the corresponding cell intrinsic transcriptional identity (Matcovitch-Natan et al., 2016). Macrophages derived from various progenitor populations (yolk sac, fetal liver, and bone marrow) appear at different stages of development, coincident with different sites of hematopoiesis. The earliest macrophages in the mouse embryo are found at embryonic day (E) 7.5, but are actually a transient population of maternally derived macrophages that are cleared by E9.0 (Bertrand et al., 2005; Kierdorf et al., 2013). Embryonic macrophages derive from the extra-embryonic yolk sac and arise from both primitive hematopoietic progenitors, which are evident as early as E7.5, as well as from erythromyeloid progenitors, which are detected by E8.5 (Bertrand et al., 2005; Schulz et al., 2012; Kierdorf et al., 2013; McGrath, Frame and Palis, 2015; Gomez Perdiguero et al., 2015; Mass et al., 2016). By E10.5, definitive hematopoietic stem cells (HSCs) are present within the aorto-gonadomesonephros (AGM) region and generate a second population of embryonic macrophages. Subsequently, AGM HSCs cells migrate to the fetal liver where they expand and differentiate starting at E12.5 (Bertrand et al., 2005; Orkin and Zon, 2008; Schulz et al., 2012; Gomez Perdiguero et al., 2015). At this point, the fetal liver is a transient source of definitive hematopoiesis from which circulating monocytes are derived. During neonatal stages, fetal liver hematopoiesis declines and is replaced by bone marrow hematopoiesis (Lichanska et al., 1999; Lichanska and Hume, 2000; Geissmann et al., 2010; Wynn, Chawla and J. W. Pollard, 2013).
Yolk-sac-derived macrophages form the first wave of tissue resident macrophages. Cell lineage tracing studies have shown that mouse yolk sac macrophages colonize the developing central nervous system (CNS) and persist to adulthood as tissue resident microglia (Ginhoux et al., 2010). These cells self-renew within the CNS throughout adulthood without significant contribution from the bone marrow and circulating blood monocytes and were shown to play important roles in the sprouting, migration, anastomosis and refinement of the CNS vascular system (Herbomel, B. Thisse and C. Thisse, 2001; Ajami et al., 2007; Ginhoux et al., 2010; Fantin et al., 2010; Hashimoto et al., 2013; Arnold and Betsholtz, 2013). For example, the microglia-deficient mice (Pu.1 knockouts and Csf1 op/op mutants) have reduced numbers of vascular branch points in the hindbrain (Fantin et al., 2010). Similarly, previous studies indicates that microglia play roles in shaping the vascular plexus in the retina (Rymo et al., 2011; Arnold and Betsholtz, 2013). However, microglia share a number of molecular characteristics with other macrophage classes, making it difficult to distinguish microglia from other macrophages using molecular marker analysis. Therefore, the detailed characterization of morphological features has been widely used as an alternative to quantitatively differentiate between microglia and other myeloid cells in culture (Fujita et al., 1996; Eder et al., 1999; Szabo and Gulya, 2013; Sheets et al., 2013).
In this study, we first investigated the possibility that EMs are endowed with greater pro-angiogenic potential than BMDMs. Despite several studies indicating that EMs express pro-angiogenic factors and gene expression signatures distinct from fetal liver and adult macrophages (Bertrand et al., 2005; Ovchinnikov, 2008; Pucci et al., 2009; Schmieder et al., 2012), very little is known about the phenotypic properties and functional capabilities of EMs before the production of the definitive HSCs. Furthermore, to date, most research investigating the pro-angiogenic potential of macrophages has been performed using adult macrophages. Therefore, isolation and characterization of EMs is necessary for determining the full potential of macrophages in driving vascular development as well as other roles served by microglia.
Here, we report successful isolation, in vitro expansion and characterization of EMs derived from E9.5 Csf1r-EGFP+/tg transgenic mice. We confirmed the macrophage identity in our isolated/cultured EMs by their expression of the specific macrophage markers Csf1r and F4/80. We also determined that EMs possess pro-angiogenic and pro-inflammatory properties distinct from adult macrophages. Most importantly, we demonstrated that EMs have a greater capability to differentiate into microglia as compared to adult macrophages when co-cultured with NSPCs, consistent with data showing that microglia derive from EMs (Ginhoux et al., 2010). The purified EM population is thus suitable to investigate a wide range of molecular properties and functional capabilities of macrophages ex vivo.
MATERIAL AND METHODS
Mice
Flk1-myr::mCherrytg/tg mice (Larina et al., 2009) were crossed to Csf1r-EGFPtg/tg mice (Sasmono et al., 2003) to generate Csf1r-EGFP+/tg; Flk1-myr::mCherry+/tg mice. Csf1r-EGF +/tg; Flk1-myr::mCherry+/tg mice were intercrossed to generate Csf1r-EGFPtg/tg; Flk1-myr::mCherrytg/tg mice. Csf1r-EGFPtg/tg males were mated to CD1 females to generate Csf1r-EGFP+/tg mice. CD1 females were purchased from the Charles River Laboratories. All animal procedures were reviewed and approved by Baylor College of Medicine Institutional Animal Care and Use Committee in compliance with the National Research Council Guide for the Care and Use of Laboratory Animals.
In vivo confocal imaging macrophages in the embryo
CD1 females were mated to Csf1r-EGFPtg/tg; Flk1-myr::mCherrytg/tg males to generate Csf1r-EGFP+/tg; Flk1-myr::mCherry+/tg transgenic embryos. The morning of a vaginal plug formation was counted as embryonic day 0.5 (E0.5). Embryos were isolated at E9.5 and dissected under a stereomicroscope at 37°C. Embryos were kept in a prewarmed dissection media (1XDMEM-F12 contains 10% Fetal Bovine Serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin Life technologies). The decidua, Reichert’s membrane, and yolk sac were removed to allow visualization of the embryo. Embryos from different litters were stage matched by somite numbers. Dissected embryos were allowed to recover for one hour in a 35 cm2 glass bottom dish containing pre-warmed dissection media at 37°C and 95% air/ 5% CO2 incubator. The dish was placed onto a Zeiss LSM 780 confocal microscope with an incubated stage (humidified at 37°C and 95% air/ 5% CO2) for imaging. Tiled-z-stack images were taken using 10X, 20X, and 40X objectives. Images were analyzed to determine the spatial distribution of EGFP+ macrophages in the embryo and their association with the vasculature (identified by mCherry expression).
Macrophage Isolation
Embryonic macrophages.
E9.5 Csf1r-EGFP+/tg transgenic embryos were harvested at room temperature (RT) and placed in cold HBSS+ (HBSS without calcium or magnesium and containing 5% FBS, 10 mM HEPES, 100 U/ml penicillin and 100 μg/ml streptomycin pH=7.4 (Life technologies)). After rapid transfer into a polypropylene tube containing cold HBSS+ stored on ice, embryos were then digested with 0.1% type I collagenase (Cat#17101015 Life technologies) for 20 minutes at 37°C in a humid incubator containing 95% air/ 5% CO2. The collagenase activity was terminated by addition of an equal volume of cold HBSS. The suspension was gently mixed by pipetting several times with a 5ml pipette and passed through a sterile 27.5 gauge needle attached to a syringe. Embryonic cell pellets were collected and resuspended in cold HBSS+ and filtered through a 40 μm cell strainer into clean tubes to generate single cell suspensions. A live cell count was determined using the 0.4% trypan blue exclusion test, and enumeration was performed with a disposable hemocytometer. Samples were centrifuged and resuspended with cold HBSS+ at a concentration of 106 cells/ml. Samples were then transferred into polypropylene fluorescence-activated cell sorting (FACS) tubes for cell sorting by EGFP signal using a BD FACSAria II cell sorter. Sorted cells were collected in polypropylene FACS tube contains DMEM+ (1X DMEM high glucose medium supplemented with 20% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin, Life technologies) and immediately cultured in a DMEM+ supplemented with 40 ng/ml macrophage colony stimulating factor (M-CSF, ProSpec) at a concentration of 5 × 105 cells/ml, and incubated in a humidified incubator at 37°C and 95% air/ 5% CO2. Csf1r-EGFP labeled EMs were used between passages 2 and 4 for further experimentations.
Bone marrow derived macrophages.
Bone marrow cells were isolated from the tibia of 10–14 week-old Csf1r-EGFP+/tg transgenic mice under sterile conditions as described previously (Kroeger, Collins and Ugozzoli, 2009; Marim et al., 2010). Briefly, tibias were placed in a 60 mm petri dish containing sterile 1XPBS, and the muscles connected to the bone were removed. In a tissue culture hood, both ends of the tibia were cut off and bone marrow cells were flushed out into a petri dish with 27.5 gauge needle attached to a 1cc syringe containing sterile 1X PBS. Bone marrow cells were homogenized and resuspended in DMEM+ media. The cell suspension was mixed thoroughly and passed through a 40 μm cell strainer to create a single cell suspension. Bone marrow cells were differentiated into BMDMs by culturing in DMEM+ supplemented with 40 ng/ml M-CSF and incubated for 4 days in a humidified incubator at 37°C and 95% air/ 5% CO2. BMDMs were selected by adhesion to petri dishes after 5 days of differentiation. Csf1r-EGFP labeled BMDMs were used between passages 2 to 4 for our experiments.
Quantitative real time PCR (qRT-PCR)
EMs or BMDMs were seeded on tissue culture treated 6-well plate at a density of 10,000 to 15,000 cells/cm2, and allowed to grow to confluence at 37°C in humidified incubator containing 95% air and 5% CO2. Confluent EMs and BMDMs were washed with cold 1XPBS and the total RNA was extracted with Trizol-reagent (Life technologies) and purified following the Qiagen RNeasy mini kit guidelines. 2μg RNA was reverse transcribed using Invitrogen SuperScript ® III cDNA synthesis kit, and 250ng cDNA was used per reaction. qRT-PCR was carried out using Taqman primers, Taqman Universal Master Mix II on a StepOnePlus Real-Time PCR system (Applied Biosystems). The following Taqman qRT-PCR primers were used: EGF-like module-containing mucin-like hormone receptor-like 1 (F4/80, Mm00802529_m1), Colony stimulating factor 1 receptor (Csf1r, Mm01266652_m1), Cluster of Differentiation Molecule 11b (CD11b, Mm00434455_m1) Inducible nitric oxide synthase (iNOS, Mm00440502_m1), Tumor Necrosis Factor alpha (TNF, Mm00443260_g1), C-C chemokine receptor type 2 (CCR2, Mm00438270-m1), Neuropilin 2 (Nrp2, Mm00803099_m1), Notch homolog 1 (Notch1, Mm00435249_m1), Arginase-1 (Arg1, Mm00475988_m1), Fractalkine receptor (CX3CR1, Mm02620111-s1) and Mannose receptor C type 1 (Mrc1, Mm01329362_m1). 18S (4333760F) level was used for normalization and relative gene expression ratios were calculated using quantitative ΔCT methods.
Immunocytochemistry
EMs or BMDMs or ANS4 were seeded at a density of 10,000 to 15,000 cells/cm2 into Lab-Tek II 8 well glass chamber slide and grown to > 80% confluence prior to staining. Cells were washed with 1XPBS for 5 minutes and fixed in 4% paraformaldehyde (PFA) for 30 minutes at RT. Cells were blocked with a blocking solution (1XPBS with 0.1% Triton-X and 2% natural serum) for 1 hour at RT, followed by incubation with primary unconjugated antibodies diluted in the blocking solution and incubated overnight at 4°C. Immunocytochemical (ICC) analyses were performed with the following primary antibodies: F4/80, Csf1r, CD11b, iNOS, TNF, CCR2, Arg1, CX3CR1, Mrc1, GFP, ionized calcium-binding adapter molecule 1 (Iba-1), Nestin, and beta class III tubulin (TUJ1) (Sup. Table1). ANS4 were incubated with Iba-1, Nestin, and TUJ1 only. Following, cells were washed 3 times with 1XPBS for 5 minutes and incubated with secondary antibodies (1:400 dilution), and 4’−6-Diamidino-2-phenylindole (DAPI) (1:500 dilution) in the blocking solution for 1 hour at RT (Sup. Table 2). After washing with 1XPBS as previously stated slides were mounted in fluoromount-G (SouthernBiotech) and images were taken using Zeiss LSM 780 confocal microscope at 20X objective. All comparative images were taken with the same laser power and gain settings in order to make qualitative comparisons between staining levels in different samples. Multiple fields of view were imaged from biological replicates and representative images are shown. The ratio of macrophage marker immunopositive cells with respect to total number of cells in a field of view was calculated using the software FARSIGHT (http://www.farsight-toolkit.org/). The total number of cells in the field of view was calculated by enumerating the number of DAPI stained nuclei using the nuclear segmentation program in FARSIGHT (Al-Kofahi et al., 2011).
Western blotting
Total cytoplasmic and membrane proteins were isolated from proliferating EMs and BMDMs cultures grown to near confluence in 100 mm tissue culture treated plates. Proteins were quantified using Bradford protein assay. 30μg of protein from each sample were denatured and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked with 5 % non-fat milk in Tris buffered saline supplemented with 0.05 % Tween-20 (TBS-T) for 1hour at RT, followed by incubation with primary unconjugated antibodies diluted in TBS-T containing 5% non-fat milk overnight at 4°C. The following primary antibodies were used: CD11b, iNOS, CCR2, Arg1, CX3CR1, Mrc1, GFP, GAPDH (Sup. Table 1). Following several washes with TBS-T, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies diluted 1:5000 in TBS-T/5% non-fat milk for 1 hour at RT (Sup. Table 2). Membranes were washed as above, treated with pierce enhanced chemiluminescence western blotting substrate plus detection reagents and visualized by generating autoradiographs following film exposure using a film processor. Protein expression levels were compared between samples and controls using spot densitometric analyses of digital autoradiographs relative to GAPDH expression using ImageJ software.
In vitro assays
Macrophage Polarization.
EMs or BMDMs were seeded at 10,000 cells/cm2 in culture medium containing 40ng/ml M-CSF onto tissue 6-well tissue culture plates and allowed to grow to > 70 confluence at 37°C in humidified incubator containing 95% air and 5% CO2. Macrophages were then polarized for 24 hours with 200ng/ml lipopolysaccharide (LPS, cat#L2630) for M1 or with 20ng/ml Interleukin-4 (IL-4, Prospec) for M2 macrophages. Non-stimulated EMs and BMDMs were used as control. Total RNA was extracted and cDNA was synthesized as previously described. Then macrophage polarization was confirmed by qRT-PCR analysis of gene expression of established M1 (CD11b, iNOS, CCR2, and TNF) and M2 (Arg1, Cx3cr1, and Mrc1) macrophage markers.
In vitro 3D-endothelial cell cord formation.
Human Umbilical Vein Endothelial Cells (HUVECs, Lonza) at a density of 2 × 106 cells/ml were encapsulated in 2.5 mg/ml collagen pre-gel solution with Csf1r-EGFP labeled EMs or Csf1r-EGFP labeled BMDMs at a density of 4 × 105 cells/ml as described before (Hsu et al., 2015). 10 μl of collagen pre-gel solution mix was gently seeded onto each well of a μ-Slide Angiogenesis (cat#81506, ibidi) and incubated at 37°C in humidified incubator containing 95% air and 5% CO2 for 30 minutes to allow the collagen pre-gel to solidify. 50 μl EGM complete medium (Lonza) supplemented with 40 ng/ml human-VEGF (ProSpec), 40 ng/ml human-FGF2 (ProSpec) and 40 ng/ml Mouse-M-CSF was added to each well and incubated in a humidified incubator at 37°C and 95% air/ 5% CO2. To test whether macrophage conditioned media could influence HUVEC tube formation, HUVECs were seeded as described above, but then were cultured with conditioned media from EMs or BMDMs, supplemented with FGF, VEGF and M-CSF as described above. To prepare conditioned media, EMs or BMDMs were plated at a density of 5×105 cells/ml in EGM complete medium. After 72 hours conditioned media were collected and stored at −20°C.
After 24 and 72 hours, collagen gels were collected from macrophage/endothelial cell co-culture experiments, washed three times with 1XPBS for 5 minutes. They were then fixed in 4 % PFA for 1 hour at RT and followed by three washes with 1XPBS. For conditioned media experiments, HUVECs within collagen gels were grown in conditioned media for 72 hours then harvested in an identical manner. Samples were then incubated in a blocking solution (1 × PBS with 0.8% Triton × and 2% donkey serum) at RT for 1 hour. Following, samples were incubated with the primary antibody platelet endothelial cell adhesion molecule (mouse anti human PECAM-1, R&D) at the ratio of 1:200 in blocking solution at 4°C overnight. Following three washes with 1XPBS, samples were incubated with Alexa donkey anti-mouse (1:400) and DAPI (1:500) diluted in blocking solution for 1 hour at RT. Samples were washed as above and mounted with fluoromount-G. Samples were examined by using a Zeiss LSM 780 confocal microscope. Z-stack images were taken using an EC Plan-Neofluar 10X/0.30 NA objective at 0.6 zoom with a step-size of 7.24μm. Since HUVECs have a diameter of 15–18μm, we assumed more than two cells would be needed to form a cord structure. Therefore, the length and number of cords over 40μm were calculated after skeletonizing vessel structures in z-stacks using a 3D open snake-tracing program in FARSIGHT (Wang et al., 2011).
Differentiation of EMs and BMDMs into Microglia.
EMs or BMDMs at a density of 1 × 105 cells/ml were co-cultured with mouse adherent neural stem cell line (ANS4) at a density of 5 × 105 cells/ml into Lab-Tek 8 well chambered coverglass using neural stem cell special culture media (contains 40ng/ml Mouse-M-CSF) as described previously (S. M. Pollard, 2013). EMs or BMDMs at a density of 1 × 105 cells/ml cultured alone in neural stem cell media were used as controls. After 3, and 20 days in culture, cells were imaged using the EGFP signal. Images were taken with a Zeiss LSM 780 confocal microscope using a 40X objective at 1.2 zoom. Binary images of macrophages were generated from the confocal images using a custom MATLAB program (MATLAB 2008a, Mathworks, Natick, MA). The program uses a multilevel thresholding method in ImageJ to create a preliminary binary image of a macrophage. The binary image is then post-processed to remove binary components not associated with the macrophage. An editable contour of the macrophage is then created in MATLAB and the contour is manually edited to fine tune the boundary of the macrophage by comparing with the original color image. Finally, the refined binary image of the macrophage is generated from the edited contour. Perimeters and areas of macrophages were calculated from their binary images. The transformation index (TI) defined as ((perimeter)2/4π × area) was calculated for each macrophage. TI reflects the degree of process extension of a macrophage (Szabo and Gulya, 2013). EMs, BMDMs and ANS4 were seeded as described above, but then after 3 and 20 days, samples were collected and immunostained with Iba-1 as described above.
Cell density measurement.
EMs or BMDMs at a density of 1 × 105 cells/ml cultured alone or with 5 × 105 cells/ml ANS4. After 1, 3, 6, 10, 13, 15, and 20 days in culture, EMs and BMDMs were imaged using the EGFP signal. Images were taken with a Zeiss LSM 780 confocal microscope using a 40X objective at 0.6 zoom. Moreover, EMs, BMDMs and ANS4 were seeded as described above, but then to visualize neural stem cells, cells were stained with and antibody against Nestin at day 3 and 20. In addition, ANS4 seeded at a density of 5 × 105 cells/ml were used as control.
Differentiation of NSPCs into neurons.
ANS4 at a density of 5 × 105 cells/ml in neural stem cell special culture media (contains 40ng/ml Mouse-M-CSF) cultured alone or with 1 × 105 cells/ml EMs or BMDMs into Lab-Tek II 8 well glass chamber slide. To examine neural differentiation, after 20 days, samples were collected and immunostained with TUJ1 as described above.
qRT-PCR.
EMs or BMDMs were seeded at a density of 6,000 cells/cm2 on the back of tissue culture treated polyester membrane transwell inserts (6.5mm diameter, 0.4μm pore size). Transwell inserts were incubated for 2 hours in a humidified incubator at 37°C and 95% air/ 5% CO2. After 2 hours, transwell inserts were placed in 24-well tissue culture plate contains 600μl neural stem cell special culture media (contains 40ng/ml Mouse-M-CSF). Then ANS4 at a density of 12,000 cells/cm2 in neural stem cell special culture media supplemented with 40ng/ml Mouse-M-CSF were cultured on the inside of tissue culture treated polyester membrane transwell inserts. At day 1 and 20, cells were collected with 0.25% Trypsin-EDTA (life technologies), washed with cold 1XPBS, a total RNA was extracted with Trizol-reagent, and purified following the Qiagen RNeasy macro kit guidelines. cDNA was synthesized as previously described. Then microglia differentiation was confirmed by qRT-PCR analysis of gene expression of Tmem119 (Mm00525305_m1). Beta-2-microglobuin ((B2M), Mm00437762_m1) level was used for normalization and relative gene expression ratios were calculated using quantitative ΔCT methods.
Statistical analysis
Statistical analyses were performed using GraphPad prism 6 software with statistical significance set at p<0.05. p value ≥0.05 (N.S. not significant), < 0.05 (*), < 0.01 (*), < 0.001(***), < 0.0001(****). All experiments were performed with an n ≥ 3. One-way ANOVA or unpaired student t-test were used to determine statistically significant differences between parametric variables based on the Shapiro-Wilk test of normality.
RESULTS AND DISCUSSION
In vivo imaging of embryonic macrophages
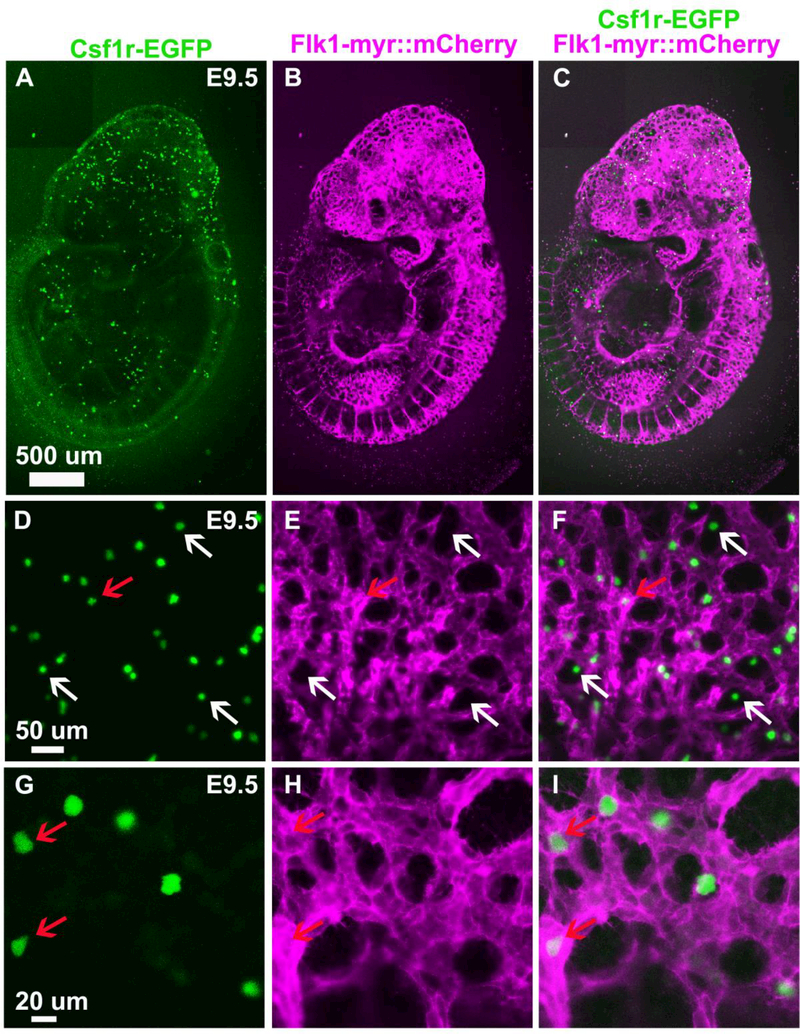
During early mouse development, EMPs and EMs express Csf1r. We have taken advantage of a Csf1r-EGFP+/tg reporter line to observe and purify EMs from the mouse embryo (Sasmono et al., 2003). First, we assessed the spatial distribution of EGFP+ cells in E9.5 Csf1r-EGFP+/tg; Flk1-myr::mCherry+/tg transgenic embryos. This time point precedes the production of definitive Hematopoietic Stem Cells (HSCs) from the aortagonad-mesonephros (AGM) region at E10.5 (Orkin and Zon, 2008; Schulz et al., 2012). Consistent with previous reports, E9.5 EGFP+ cells were found dispersed throughout the embryo with greater density anteriorly (Fig. 1 A, C), and appeared to be in close association with the Flk1-myr::mCherry+ vasculature (Fig 1. B, E, H) (Bertrand et al., 2005; Ginhoux et al., 2010; Schulz et al., 2012). High magnification imaging revealed Csf1r-EGFP+ cells in both the lumen of the vasculature and in extravascular spaces (Fig. 1 F, and I). At this stage, the cells exhibited a more rounded appearance and lacked the branched processes and extensions (Fig 1. D, G), similar to other studies describing yolk-sac-derived macrophages during early mouse embryogenesis (DeFalco et al., 2014). These early macrophages are likely yolk sac derived since they appeared prior to the formation of definitive HSCs in the AGM region but after the onset of circulation (Orkin and Zon, 2008; Ginhoux et al., 2010; Schulz et al., 2012). The anterior bias of macrophages is consistent with observations made by others that yolk sacderived macrophages move through the circulation to become microglia (Arnold and Betsholtz, 2013).
Figure 1.
Csf1r-EGFP+ cells are found in and around the vasculature (Flk1-myr::mCherry+). A., B. and C. Confocal images of an E9.5 Csf1r-EGFP+/tg; Flk1-myr::mCherry+/tg transgenic embryo showing EGFP+ embryonic macrophages dispersed throughout the embryo with greater density anteriorly. D. to I. High magnification images of the anterior dorsal part of the embryonic head at E9.5. EGFP+ embryonic macrophages appear in close association with the vasculature, in both the lumen of the vasculature (red arrow) and extravascular spaces (white arrow), and exhibit a rounded morphology.
Primary cultures of Csf1r-EGFP +/tg embryo-derived EMs
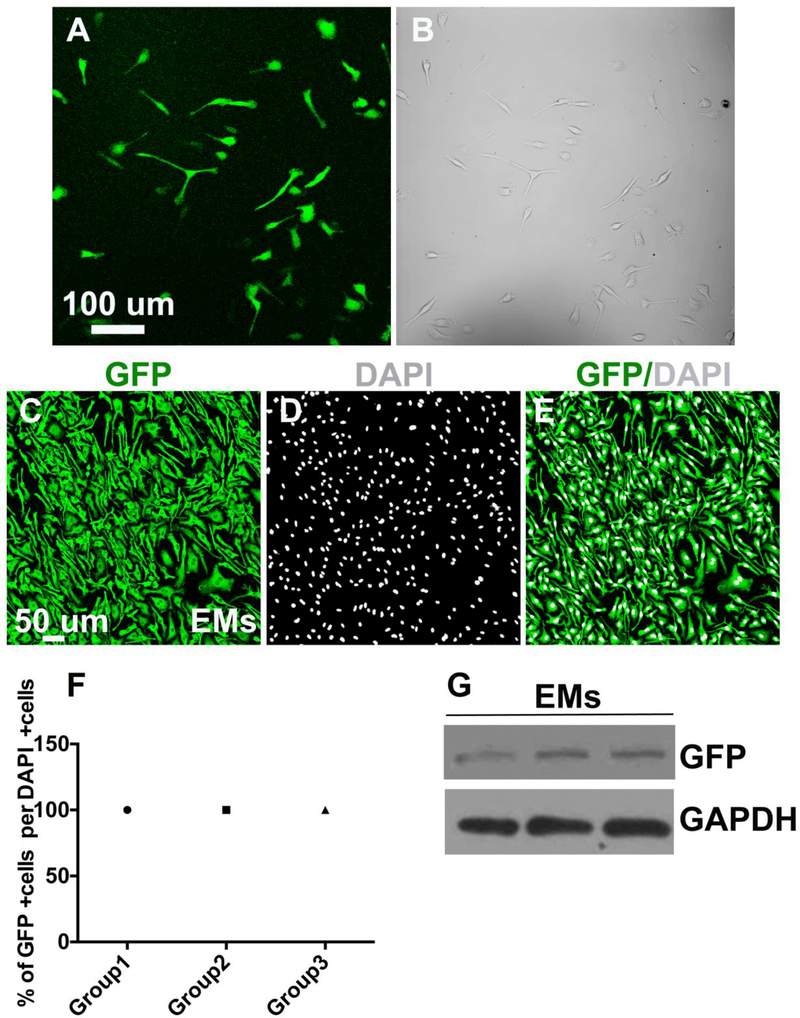
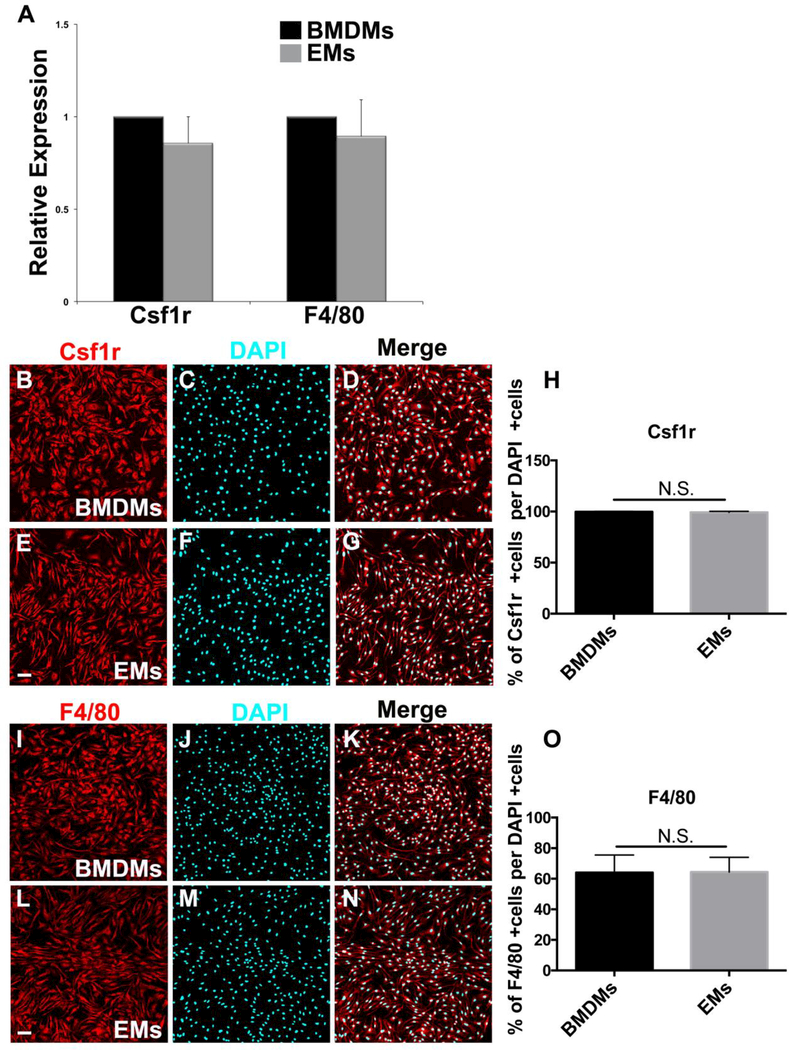
To obtain pure isolates of EMs, E9.5 Csf1r-EGFP embryos were enzymatically digested and EGFP+ cells were isolated using FACS. The Csf1r-EGFP transgene has been shown to recapitulate the endogenous expression patterns of Csf1r. Csf1r mRNA is detectable in the earliest yolk sac macrophages and the expression in the embryo and adult mouse is largely restricted to cells of macrophage lineage (Sasmono et al., 2003). Despite the low density of EGFP+ macrophages in E9.5 embryo, we successfully isolated EMs from E9.5 Csf1r-EGFP embryos. After enzymatic digestion of embryonic tissues, EGFP+ cells were isolated by FACS then cultured. Freshly sorted Csf1r-EGFP+ EMs were obtained with a very high purity and, within 24 hours of plating, formed a monolayer and displayed a macrophage-like morphology (Fig. 2A). As illustrated in Fig. 2B, bright field (BF) imaging showed that all sorted cells were EGFP+. The EGFP signal in sorted EMs was confirmed by GFP antibody labeling (Fig. 2 C-E) and western blot analysis (Fig. 2G). Quantification from merged images of GFP and DAPI+ nuclei (Fig. 2 E) showed that 100% of DAPI+ EMs were GFP+ (Fig. 2F). There were no cells that were DAPI+ but did not express GFP, which confirmed that the flow cytometric isolation of EMs by EGFP selection yielded a highly pure EM population. The macrophage identity of EMs isolates was further supported by examining the expression Csf1r and F4/80, which are well characterized mouse macrophage markers and extensively used to detect macrophages in the yolk sac, embryo and adult mouse (Lichanska et al., 1999; Lichanska and Hume, 2000; Schulz et al., 2012). We compared these results to similar experiments performed on bone marrow-derived macrophages (BMDMs) sorted from Csf1r-EGFP+/tg adult transgenic mice. EM expression levels of Csf1r and F4/80 mRNA showed no statistically significant differences compared to BMDMs (Fig. 3A). Consistent with the qRT-PCR data, immunofluorescence showed that there was no significant difference in Csfr1 or F4/80 immunofluorescence or staining distribution (Fig 3B-O). Thus, flow cytometric cell sorting from E9.5 Csf1r-EGFP+/tg embryos allows for the selective isolation of EMs.
Figure 2.
Isolation of a pure EGFP-expressing EM population from Csf1r-EGFP+/tg transgenic embryos A. Confocal image of sorted fluorescent EMs in culture showing their macrophage-like morphology and monolayer growth B. Phase contrast image of cultured EMs confirming that monolayers contain only fluorescent cells. C-G. Fluorescent isolated cells are EGFP-expressing EMs: C. Fluorescence ICC confocal image of EMs stained with GFP antibody (green) D. EMs stained with nuclear counterstain DAPI (gray) E. Merged image of GFP antibody labeling and DAPI F. Quantification of the GFP and DAPI signals demonstrating that 100% of DAPI positive EMs were GFP positive. G. Immunoblot image of GFP showing that EMs express GFP at the protein level. N = 3 groups. Scale bars 50 μm
Figure 3.
Macrophage identity of E9.5 Csf1r-EGFP+/tg FACS cells. A. Csf1r and F4/80 mRNA expression showed no significant differences between EMs and BMDMs. B-D. Fluorescence ICC images of BMDMs stained with Csf1r (red) and DAPI (blue). E-G. Fluorescence ICC images of EMs stained with Csf1r (red) and DAPI (blue). H. Quantification of the Csf1r signal demonstrating that ~100% of DAPI positive BMDMs and EMs were CSf1r positive. I-K. Fluorescence ICC images of BMDMs stained with F4/80 (red) and DAPI (blue). L-N. Fluorescence ICC images of EMs stained with F4/80 (red) and DAPI (blue). O. Quantification of the F4/80 signal demonstrating ~70% of DAPI positive BMDMs and EMs were F4/80 positive. Error bars indicate SE of the mean. N = 3. Scale bars 50 μm.
Reduction of pro-inflammatory gene/protein expression signature in EMs as compared to BMDMs
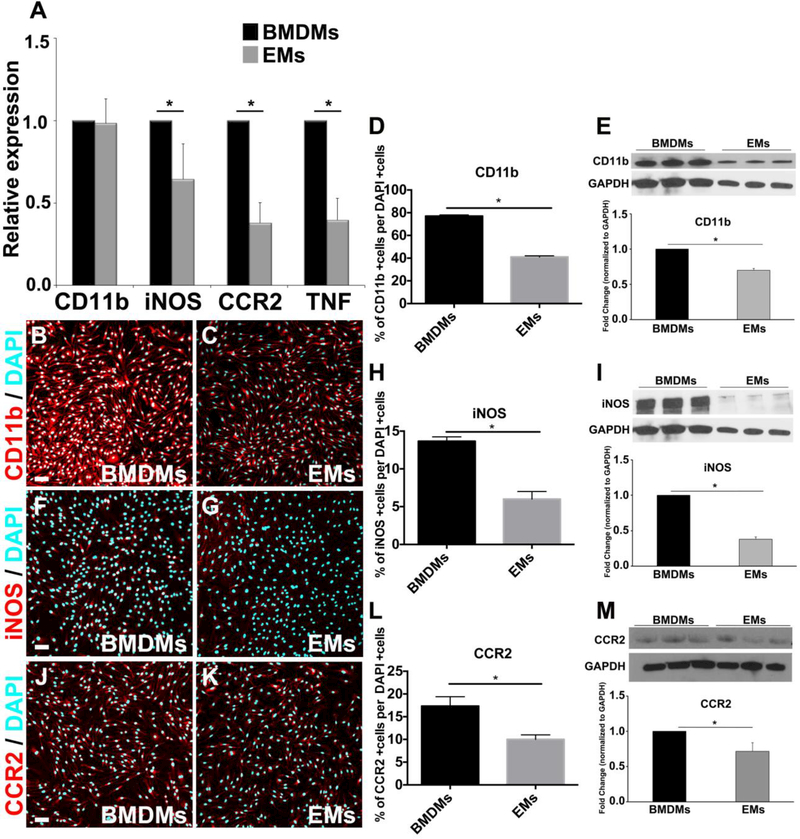
A few studies have suggested that EMs have gene signatures distinct from adult macrophages (Lichanska et al., 1999; Lichanska and Hume, 2000). Thus, we asked how cultured EMs differed from isolated BMDMs. Using qRT-PCR, we compared the expression levels of the transcripts of pro-inflammatory macrophage markers including CD11b, iNOS, CCR2 and TNF alpha, which are known to be up-regulated on activated macrophages during inflammation (Willenborg et al., 2012; Duan et al., 2016; Eisenman et al., 2017). We found that EMs expressed significantly less iNOS, CCR2, and TNF alpha mRNA compared to BMDMs (Fig. 4A). While CD11b mRNA levels were similar between cell types, immunofluorescent and Western blot analysis of CD11b did indicate a reduction in protein expression in EMs relative to BMDMs (Fig. 4B-E and Supp. Fig. 1). Consistent with this observation, only a smaller percentage of EMs were labeled with iNOS and CCR2 antibodies and Western blot showed an overall reduction in iNOS and CCR2 protein level in EMs (Fig. 4F-M and Supp. Fig. 1). It is important to note that we did not observe significant differences in TNF alpha protein expression between EMs and BMDMs (Supp. Fig. 2). While TNF alpha expression has been reported to increase during macrophage inflammatory response (Mantovani et al., 2004), it is also a signaling molecule that plays an important role in negatively regulating proliferation during development (Wride and Sanders, 1995; Kawamura et al., 2007). Therefore it is possible that EMs might use TNF alpha as a developmental cue to control cell proliferation during development. Taken together, these results suggest that EMs exhibit a less pro-inflammatory phenotype as compared to BMDMs, which is in line with previous reports on yolk-sac-derived macrophages (Schulz et al., 2012; Mizutani et al., 2012).
Figure 4.
Expression of pro-inflammatory macrophage markers by EMs A. Comparative transcript expression analysis of pro-inflammatory macrophage markers, CD11b, iNOS, CCR2, and TNF in EMs and BMDMs. EMs significantly expressed less iNOS, CCR2, and TNF than BMDMs, while CD11b expression was similar between EMs and BMDMs. B. and C. Merged fluorescence ICC images of BMDMs (B) and EMs (C) stained with CD11b (red) and DAPI (blue). D. Quantification analysis of the CD11b ICC images using Farsight toolkit demonstrating that significantly fewer EMs express CD11b compared to BMDMs. E. Immunoblot and semi-quantitative analysis of CD11b showed that EMs significantly expressed less CD11b compared to BMDMs. F. and G. Merged fluorescence ICC images of BMDMs (F) and EMs (G) stained with iNOS (red) and DAPI (blue). H. Quantification analysis of the iNOS ICC images using Farsight toolkit showed that significantly fewer EMs express iNOS compared to BMDMs. I. Immunoblot and semi-quantitative analysis of iNOS showed that EMs significantly expressed less iNOS compared to BMDMs. J. and K. Merged fluorescence ICC images of BMDMs (J) and EMs (K) stained with CCR2 (red) and DAPI (blue). L. Quantification analysis of the CCR2 ICC images using Farsight toolkit showed that significantly fewer EMs express CCR2 compared to BMDMs. M. Immunoblot and semi-quantitative analysis of CCR2 showed that EMs significantly expressed less CCR2 compared to BMDMs. Error bars indicate SE of the mean. N = 3. Scale bars 50 μm.
Amplified pro-angiogenic gene signature in EMs as compared to BMDMs
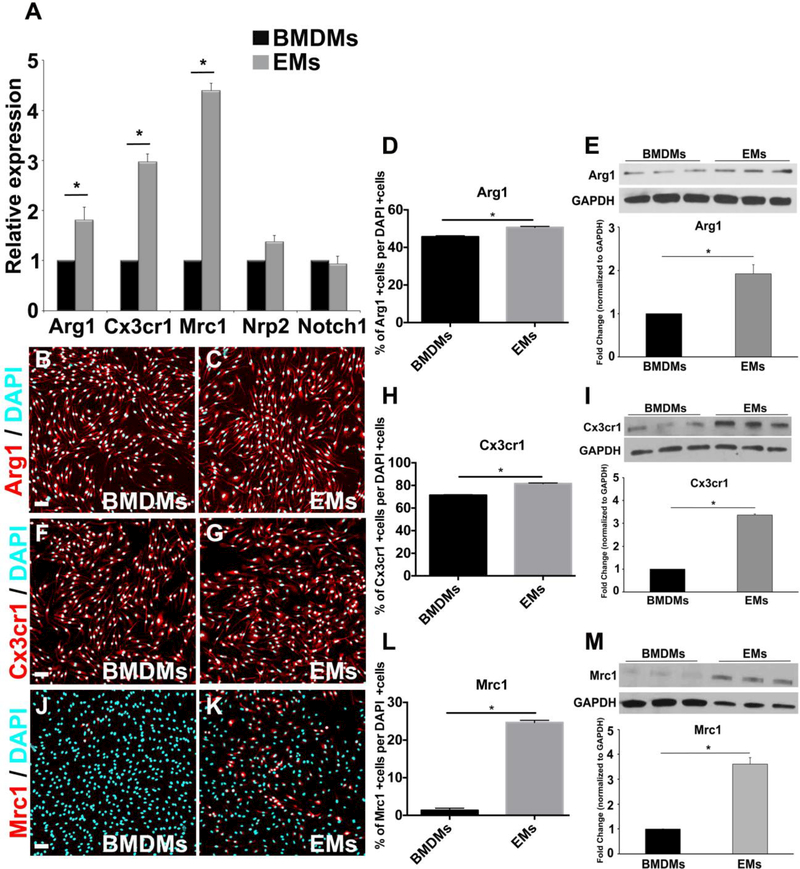
Next, we compared the mRNA expression levels of the pro-angiogenic markers Arg1, Cx3cr1, Mrc1, Nrp2 and Notch1 between EMs and BMDMs. We determined that EMs expressed significantly higher levels of Arg1, Cx3cr1, and Mrc1 mRNA compared to BMDMs, while both EMs and BMDMs equivalently expressed Nrp2 and Notch1 (Fig. 5A). Consistently, immunofluorescence showed that EM cultures contained more Arg1, Cx3cr1, and Mrc1 positive cells than BMDM cultures (Fig 5B-D, F-H, J-L and Supp. Fig. 1), This was especially true for Mrc1, which was scarcely detected in BMDM cultures, while more than 20% of the EMs were brightly positive for this marker. Western blot analysis also indicated higher levels of Arg1, Cx3cr1, and Mrc1 (Fig 5E, I and M), while no statistically significant differences were observed with the Nrp2 and Notch1 protein expression (data not shown). Taken together, these data show that EMs express higher level of pro-angiogenic markers than BMDMs, suggesting that EMs may have greater pro-angiogenic properties than adult macrophages. This is in line with previous observations that increased in Arg1 and Mrc1 expression strongly associates with the potent pro-angiogenic activities of M2 and tumor associated macrophages (TAMs) (Leitinger and Hohenester, 2007; Ho and Sly, 2009; Madsen and Bugge, 2013; Madsen et al., 2013). The high expression of Cx3cr1 by EMs moreover provides evidence of their yolk sac origin. Cx3cr1 is known to be highly expressed in yolk sac macrophage precursors in early mouse embryos and has been shown to distinguish yolk-sac-derived macrophages from monocyte-derived macrophages (Mizutani et al., 2012). Altogether our data show that EMs have unique gene signatures compared to BMDMs, which is in agreement with previous reports on these classes of macrophages (Lichanska and Hume, 2000; Pucci et al., 2009; Schulz et al., 2012). Yet, it is not clear which factors are required for macrophage-specific functions in these cells, this will need to be determined in future studies.
Figure 5.
Expression of pro-angiogenic macrophage markers by EMs A. Comparative transcript expression analysis of pro-angiogenic macrophage markers, Arg1, Cx3cr1, Mrc1, Nrp2 and Notch1 in EMs and BMDMs. EMs significantly expressed more Arg1, Cx3cr1 and Mrc1, while both EMs and BMDMs equivalently expressed Nrp2 and Notch1. B., and C. Merged fluorescence ICC images of BMDMs (B) and EMs (C) stained with Arg1 (red) and DAPI (blue). D. Quantification analysis of the Arg1 ICC images using Farsight toolkit showed that significantly more EMs express Arg1 compared to BMDMs. E. Immunoblot and semi-quantitative analysis of Arg1 showed that EMs significantly expressed more Arg1 compared to BMDMs. F. and G. Merged fluorescence ICC images of BMDMs (F) and EMs (G) stained with Cx3cr1 (red) and DAPI (blue). H. Quantification analysis of the Cx3cr1 ICC images using Farsight toolkit showed that significantly more EMs express Cx3cr1 compared to BMDMs. I. Immunoblot and semi-quantitative analysis of Cx3cr1 showed that EMs significantly expressed more Cx3cr1 compared to BMDMs. J. and K. Merged fluorescence ICC images of BMDMs (J) and EMs (K) stained with Mrc1 (red) and DAPI (blue). L. Quantification analysis of the Mrc1 ICC images using Farsight toolkit showed that significantly more EMs express Mrc1 compared to BMDMs. M. Immunoblot and semiquantitative analysis of Mrc1 showed that EMs significantly expressed more Mrc1 compared to BMDMs. Error bars indicate SE of the mean. N = 3. Scale bars 50 μm.
EMs can be polarized to both M1 and M2 phenotypes
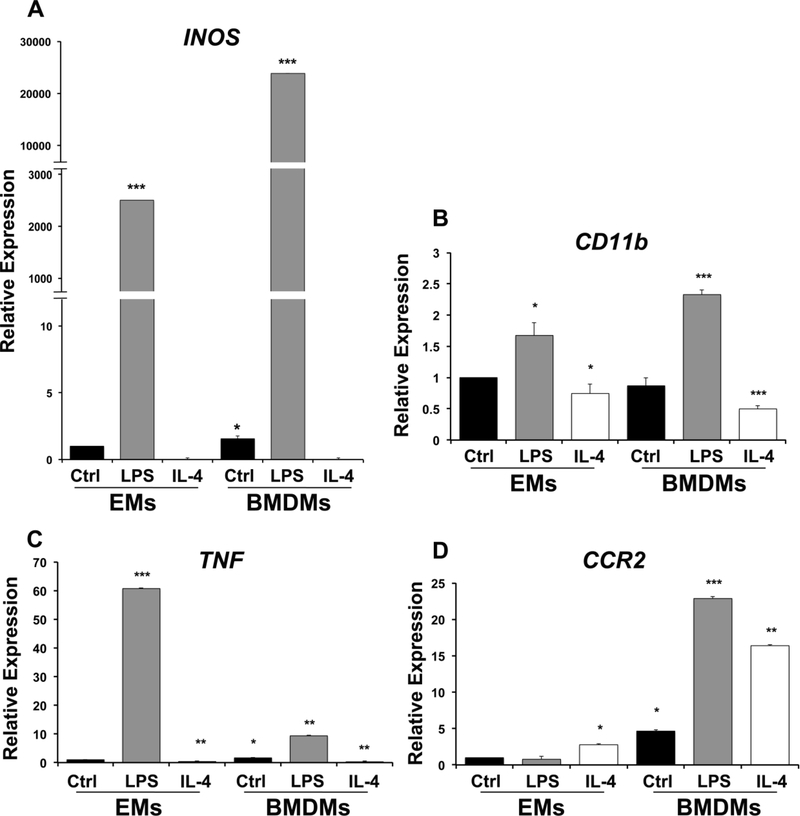
Although it is known that EMs are more likely to express pro-angiogenic (M2) markers than adult macrophages (Ovchinnikov, 2008; Pucci et al., 2009; Mazzieri et al., 2011), we sought to determine if EMs have a similar capacity as BMDMs to become polarized toward both the M1 and M2 phenotypes. EMs and BMDMs were treated with the M1 stimulus lipopolysaccharide (LPS) and M2 stimulus Interleukin-4 (IL-4). This was followed by the assessment of the levels of established M1 (iNOS, CD11b, CCR2, and TNF alpha) and M2 markers (Arg1, Cx3cr1, and Mrc1). We determined that EMs and BMDMs behaved similarly upon treatment with LPS or IL4 for both pro-inflammatory (Fig. 6) and pro-angiogenic (Fig. 7) markers with a few notable exceptions. Upon exposure to either LPS or IL4, both EMs and BMDMs showed similar changes in mRNA expression of iNOS, CD11b, and TNF alpha in response to both signaling factors as compared to controls (Fig. 6A, B, C). However, while CCR2 mRNA expression was upregulated by both LPS and IL4 in BMDMs, EMs showed only a modest, but significant increase in CCR2 in response to IL4 but no change in expression when treated with LPS (Fig. D). This is consistent with previous reports showing that yolk sac progenitors express only low levels of CCR2 (CCR2low) (Epelman et al., 2014; Liu et al., 2017).
Figure 6.
Expression profiles of pro-inflammatory macrophage markers in LPStreated/M1 and IL-4 treated/M2 macrophages A. iNOS expression was significantly upregulated in untreated BMDMs and LPS treated EMs/BMDMs and attenuated in IL-4 treated EMs /BMDMs as compared to untreated EMs. B. CD11b expression was significantly up-regulated in LPS treated EMs/BMDMs and attenuated in IL-4 treated EMs/BMDMs as compared to untreated EMs. C. TNF expression was significantly upregulated in untreated BMDMs, LPS treated EMs/BMDMs and attenuated in IL-4 treated EMs/BMDMs as compared to untreated EMs. D. CCR2 expression was significantly upregulated in IL-4 treated EMs, untreated BMDMs, LPS and IL-4 treated BMDMs, but no statistically significant differences were observed in LPS treated EMs compared to untreated EMss. Error bars indicate SE of the mean. N=3.
Figure 7.
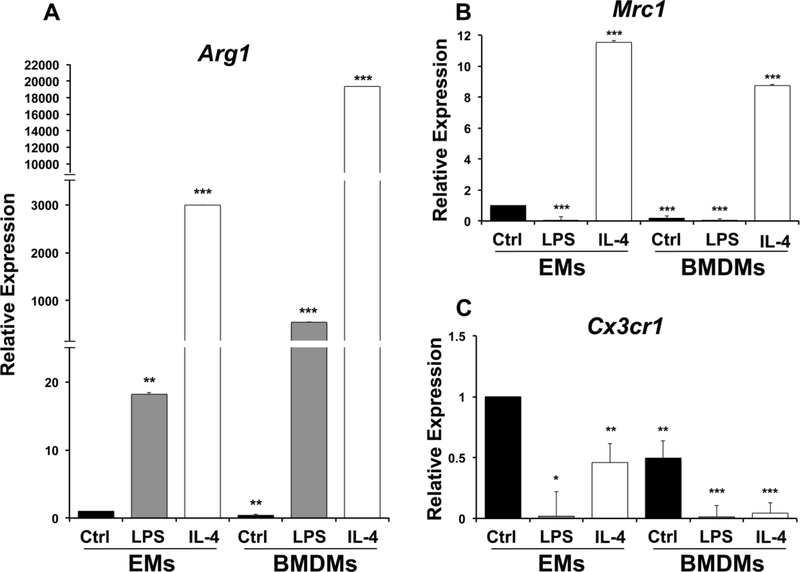
Expression profiles of pro-angiogenic macrophage markers LPS-treated or M1 driven macrophages and IL-4 treated or M2 driven macrophages A. EMs significantly express more of Arg1 as compared to untreated BMDMs. IL-4 and LPS treated EMs/BMDMs significantly express more Arg1 as compared to untreated EMs. B. Mrc1 expression was significantly attenuated in LPS treated EMs, BMDMs and LPS treated BMDMs and up-regulated in IL-4 treated EMs /BMDMs as compared to untreated EMs. C. Cx3cr1 expression was significantly attenuated in BMDMs, IL-4 and LPS treated EMs/BMDMs as compared to untreated EMs. Error bars indicate SE of the mean. N=3.
When mRNA expression of pro-angiogenic (M2) markers was examined, similar responses were observed in EMs and BMDMs for Arg1 and Mrc1 (Fig. 7A, B), but untreated EMs expressed higher levels of Cx3cr1 than untreated BMDMs and while treatment with LPS or IL4 greatly reduced expression of Cx3cr1 in BMDMs down to near undetectable levels, EMs still maintained significant expression of this marker in the presence of IL4 (Fig. 7C). Cx3cr1 has been shown to be expressed by EMs in the yolk sac and Cx3cr1+ cells have been shown to be microglial progenitors (Ginhoux et al., 2010; Mizutani et al., 2012). Moreover, Pinto, et al. has also shown that Cx3cr1 gene expression is lost with age in a subtype of cardiac tissue macrophages. This age-related loss of Cx3cr1 in cardiac tissue macrophages may relate to their exposure to inflammatory cytokines in adult hearts (Pinto et al., 2014). However, these data might suggest that reduced Cx3cr1 expression with age may result from replacement of tissue-resident macrophages, derived from EMs, with macrophages derived from the bone marrow. Overall, these data show the EMs can certainly be influenced by factors that can polarize BMDMs, but also show differences in the expression of markers such as CCR2 and Cx3cr1 that may reflect their embryonic origins.
EMs promote cord formation in vitro
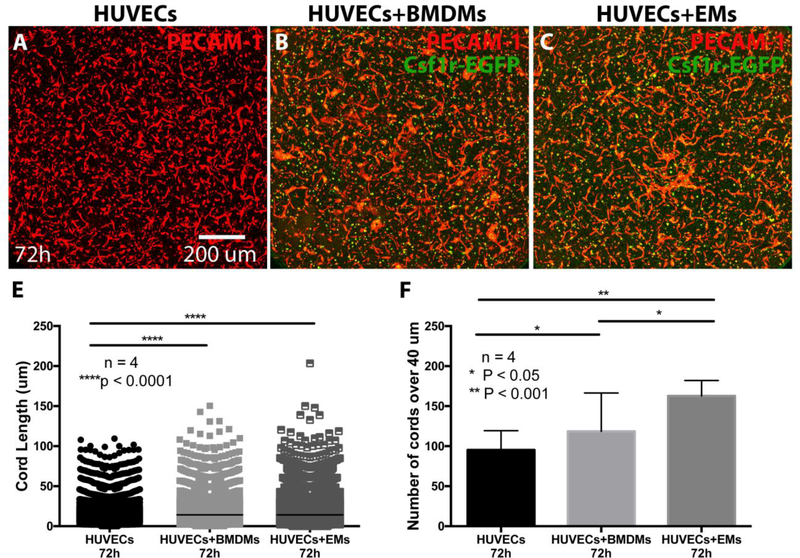
Our lab recently showed that BMDMs promote the formation of vessels in vivo and in vitro (Hsu et al., 2015). Here, we compared the pro-angiogenic potential of EMs vs. BMDMs using a 3D in vitro cord formation assay. We co-cultured HUVECs with Csf1r-EGFP labeled EMs or BMDMs at the ratio of 5:1 in collagen gels in the presence of VEGF, FGF2 and M-CSF. After 24 and 72 hours, we analyzed the ability of EMs or BMDMs to promote the formation of vascular structures. PECAM-1 immunofluorescence staining was used to visualize HUVECs and to quantify the number and length of angiogenic cords. After 24 hours, we observed that HUVECs formed cords, while the addition of EMs or BMDMs did not enhance the number or length of cords compared to the control (data not shown). Interestingly, after 72 hours, we observed that HUVECs co-cultured with BMDMs (Fig. 8B) or with EMs (Fig. 8C) exhibited an increased number and length of cord structures as compared to the control (Fig. 8A). Quantification using the 3D open snake-tracing program in FARSIGHT showed that the addition of EMs or BMDMs to HUVECs significantly increased the length of the cord structures over 40 μm as compared to HUVECs cultured alone, but no statistically significant differences were observed between EMs/HUVECs and BMDMs/HUVECs co-cultures (Fig. 8E). However, we found that the addition of EMs to HUVECs significantly increased the number of cords over 40 μm as compared to HUVECs co-cultured with BMDMs or cultured alone (Fig. 8F).
Figure 8.
EMs increased cord length and numbers in a 3D collagen base in vitro system. After 72 hours, the cord strustures formed by HUVECs cultured alone (A), HUVECs co-cultured with BMDMs (B), and HUVECs co-cultured with EMs (C) imunnostained with PECAM-1 and imaged with confocal microscopy. In the EMs/HUVECs and BMDM/HUVECs co-cultures the distribution of the cord lengths over 40 μm (E) were significantly increased compared to HUVECs cultured alone. In EMs/HUVECs co-cultures the numbers of cords over 40 μm were significantly increased compared to HUVECs co-cultured with BMDMs or alone (F).
The in vitro cord formation assay does not allow for discrimination of the effect of cell-cell interactions from that of secreted factors that diffuse into the media. To investigate this, gel encapsulated HUVECs were incubated with conditioned medium from EM or BMDM cultures grown for 72 hours. The control group was incubated with fresh EGM complete medium. After 72 hours, PECAM-1 immunofluorescence staining was used to visualize HUVECs. We observed that HUVECs in all three conditions formed cords, but HUVECs incubated with EM conditioned media formed fewer cords compared to HUVECs incubated with BMDM conditioned media and control (Supp. Fig. 3). These data show that diffusible molecules are not responsible for the increased cord formation promoted by co-culture with EMs. Thus, it is likely that direct contact or close proximity is needed between EMs and ECs to promote angiogenesis. These data agree with previous studies that have reported the importance of yolk sac macrophages in facilitating vascular anastomosis by acting as cellular chaperones to mediate the fusion of endothelial tip cells (Fantin et al., 2010; Rymo et al., 2011).
EMs differentiate into microglia in the presence of NSPCs, in vitro
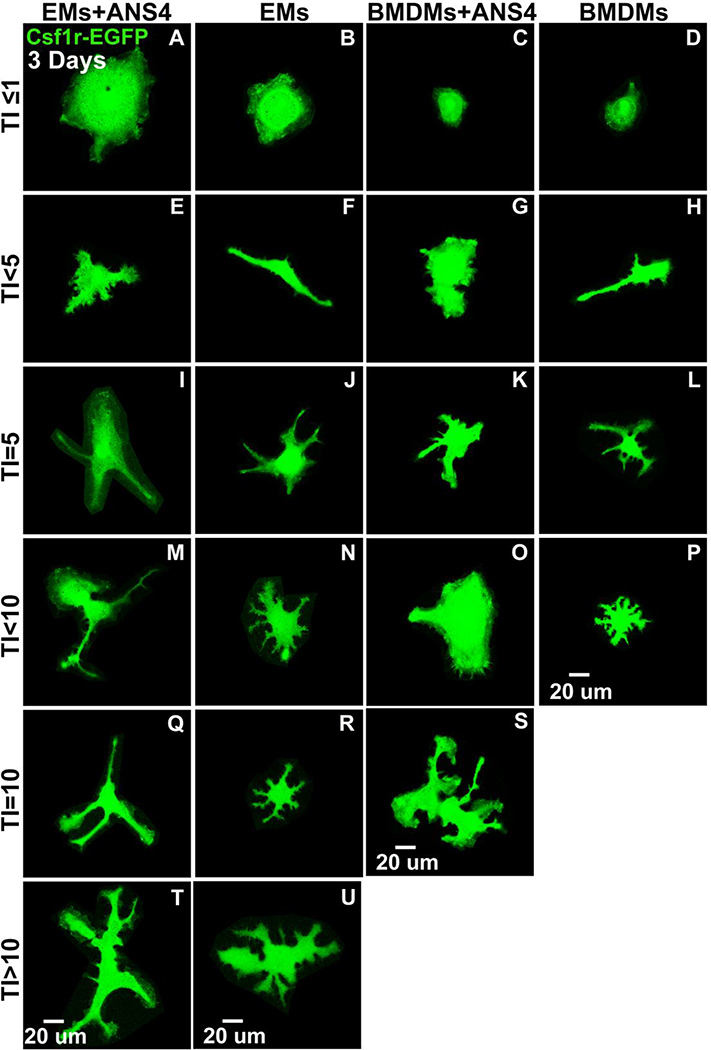
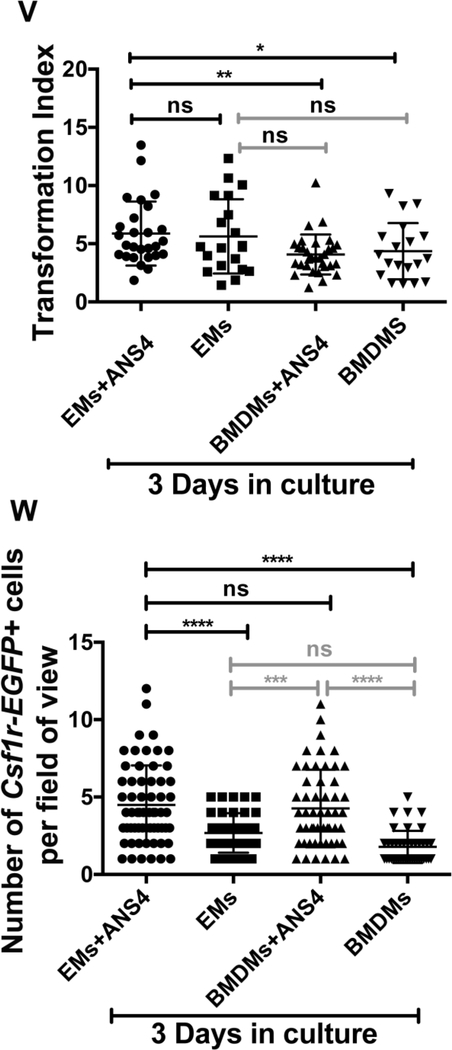
Recent evidence has shown that microglia of the CNS, are derived from yolk sac macrophages that travel through the circulation, as soon as it starts, to populate the early embryonic brain (Arnold and Betsholtz, 2013). These cells exit the yolk sac as rounded cells, but after several days within the embryonic CNS, they develop a highly ramified morphology with many branches, which is typical of microglia. First, we compared whether EMs or BMDMs had the capacity to differentiate into microglia. We found that both EMs and BMDMs did not differentiate and displayed elongated forms when cultured for 22 days (Supp. Fig. 4). Then, we tested whether cultured EMs had the capacity to form microglia in the presence of neural cells. EMs and BMDMs were co-cultured with ANS4 neural stem/progenitor cells (NSPCs) or alone. ANS4 cells were previously shown to proliferate well in culture, maintain a pluripotent state, and differentiate into neurons upon addition of growth factors (S. M. Pollard, 2013). After 3 days and 20 days, EGFP+ cells were imaged and their morphological changes were recorded and quantified by a transformation index (TI). The TI reflects the degree of morphological differentiation or process extension of a cell into the highly ramified morphology indicative of microglia (Szabo and Gulya, 2013). After 3 days, EGFP+ cells had mixed morphologies in all culture conditions (Fig. 9A-U), including many rounded, elongated and amoeboid forms with TI values <10 (Fig. 9A-P). At this early time point, a few ramified cells with TI≥10 to TI<15 were observed in the cultures of EMs alone, as well as in the EMs/ANS4, and BMDMs/ANS4 co-cultures (Fig. 9Q-U), but were not found in BMDMs cultured alone. Quantitative analysis showed a significant increase in the TI value in EMs/ANS4 co-cultures as compared to the other culture conditions (Fig. 9V). We also observed a higher density of EMs and BMDMs in the groups that contained NSPCs suggesting that these cells may provide survival or proliferation signals to EMs and BMDMs. We determined that the numbers of EMs per field of view were significantly higher in EMs/ANS4 co-cultures as compared to the other conditions (Fig. 9W).
Figure 9.
After 3 days, EMs differentiated into amoeboid/ramified microglia morphologies in the presence of NSPCs. Csf1r-EGFP transgene was used to visualize the morphological changes of EMs and/or BMDMs into microglial phenotypes. (A, B, C, D, E, F, G and H) Confocal images of EMs + ANS4, EMs, BMDMs + ANS4 and BMDMs with TI ≤ 5 displayed rounded and elongated morphologies. (I, J, K, L, M, N, O and P) Confocal images of EMs + ANS4, EMs, BMDMs + ANS4 and BMDMs with TI < 10 displayed more amoeboid forms. (Q, R, S, T and U) Confocal images of EMs + ANS4, EMs and BMDMs + ANS4 with TI ≥ 10, displayed more ramified cell forms, but not BMDMs cultured alone. (V) Quantitative analysis of the morphological changes of EMs or BMDMs into microglia as shown in the distribution of the TI values as function of the culture conditions, the TI values of EMs + ANS4 significantly increased compared to the TI values of BMDMs + ANS4 and BMDMs cultured alone, but no statistically significant differences were observed between EMs + ANS4 and EMs cultured alone. (W) Density measurements in EMs + ANS4, EMs, BMDMs + ANS4 and BMDMs cultures. The data show significantly a higher density of EMs identified in groups that contained NSPCs compared to the other groups.
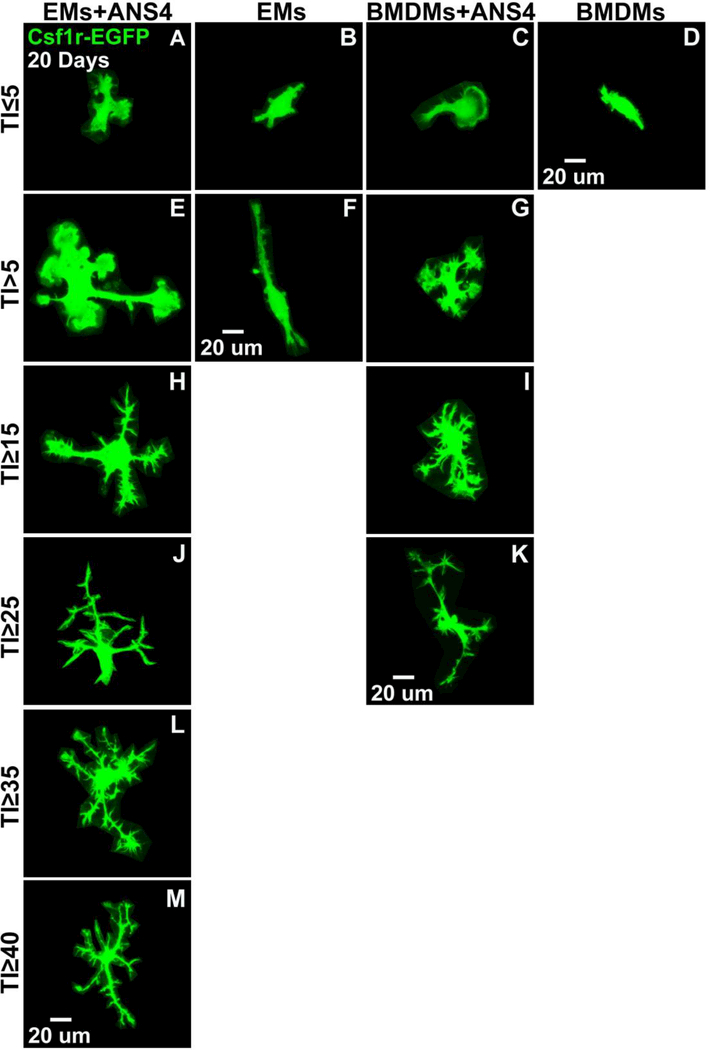
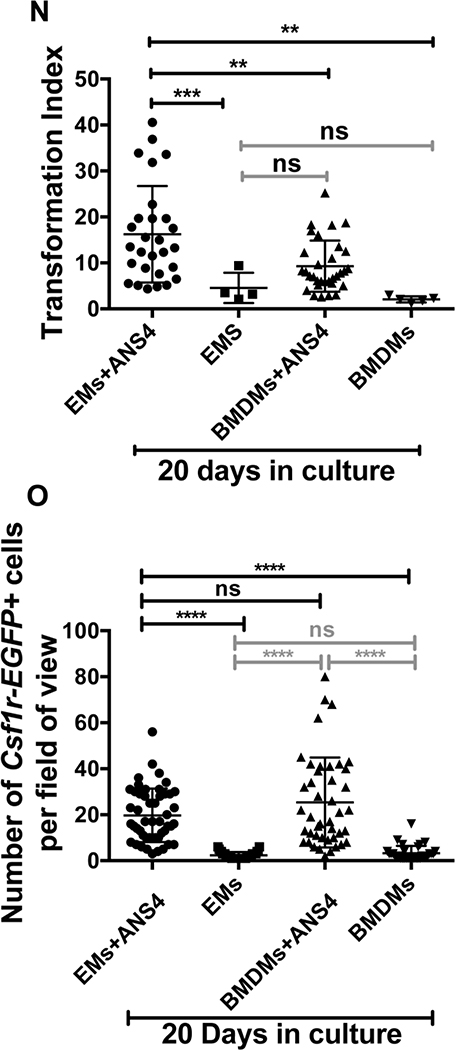
After 20 days, mixed morphological forms of EMs/BMDMs were present in all culture conditions (Fig. 10A-M). A few cells displayed elongated and amoeboid forms, with little ramification (lowest group with TI values ≤ 5) (Fig. 10A-G). Slightly ramified cells (with TI≥25 to TI<35) were only observed in EMs/ANS4, and BMDMs/ANS4 cocultures (Fig. 10H-K), but not in EMs or BMDMs cultured alone. We also found that only EMs co-cultured with ANS4 differentiated into extensively ramified morphologies with TI≥ 35 to TI<50 (Fig. 10L-M). Quantitative analysis based on the TI value showed that the degree of morphological differentiation into more microglial ramified morphology was significantly increased in EMs/ANS4 co-cultures as compared to BMDMs/ANS4 cocultures, EMs and BMDMs cultured alone (Fig. 10N).
Figure 10.
After 20 days, EMs differentiated into more ramified microglia morphologies in the presence of NSPCs. (A, B, C and D) Confocal images of EMs + ANS4, EMs, BMDMs + ANS4 and BMDMs with TI < 5 displayed rounded and elongated cell forms. (E, F and G) TI > 5, EMs + ANS4 and BMDMs + ANS4 displayed more amoeboid forms, no BMDMs cultured alone were observed with TI > 5. (H, I, J and K) TI > 15, EMs + ANS4 and BMDMs + ANS4 displayed ramified cell forms, but no EMs cultured alone were observed at TI values > 15. (L and M) Only EMs + ANS4 were observed with TI > 35 and they displayed more ramified cell forms. (N) Quantitative analysis of the morphological changes of EMs or BMDMs into microglia as shown in the distribution of the TI values as function of the culture conditions, EMs + ANS4 significantly had higher TI values compared to the other conditions. (O) Density measurements in EMs + ANS4, EMs, BMDMs + ANS4 and BMDMs cultures. The data show a higher density of EMs or BMDMs identified in groups that contained NSPCs indicating that NSPCs may support the survival or proliferation of EMs and BMDMs.
A recent study suggested that the trans-membrane protein 119 (Tmem119), as a highly specific marker of mature microglia in both mouse and human (Bennett et al., 2016). To test molecularly whether EMs vs. BMDMs formed into bona fide microglia, we examined the mRNA expression level of Tmem119 between EMs, EMs+ANS4, BMDMs, BMDMs+ANS4, and ANS4 cultured under the influence of neural environment. At day 1, Tmem119 mRNA was not detectable in either EMs or BMDMs cultured alone or with ANS4. Interestingly, at day 20, both EMs and EMs+ANS4 expressed Tmem119, but EMs+ANS4 significantly expressed more Tmem119 compared to EMs cultured alone. In contrast, we found that Tmem119 was not detectable in BMDMs, BMDMs+ANS4, and ANS4 (Supp. Fig. 5), in agreement with previous findings that microglia derived from bone marrow (BM) progenitors do not express Tmem119 and engrafted BM cells retain their non-microglial identity, even after 6 months in the adult CNS (Bennett et al., 2016). Interestingly, the level of Tmem119 expression paralleled the findings from morphological analysis. Tmem119 expression was increased in EMs+ANS4 and we also showed that EMs preferentially differentiated into microglial ramified morphology when co-cultured with ANS4 for 20 days. The expression studies also indicate that EMs cultured alone can express this marker and we have detected a small number of cells with a lower transformation index in these cultures (Fig. 9U and Fig. 10F) suggesting that Tmem119 expression is tightly regulated by the degree of EMs differentiation into mature microglia. These data are in line with previous in vivo findings that Tmem119 expression is developmentally regulated and correlates with microglia maturity. Prenatal immature microglia in mice express a low level of Tmem119, but the expression is highly increased in mature microglia postnatally (Bennett et al., 2016). These observations raise important questions for future studies: What signals do the ANS4 cells produce at day 20 that enhanced microglia maturation? Does ANS4 differentiation, similarly to CNS maturation, induce a signal that acts on microglia?
We also analyzed the protein expression of the microglia marker ionized calciumbinding adaptor molecule 1 (Iba-1). First, we compared the protein expression levels of Iba-1 between EMs, BMDMs, and ANS4 cultured alone in their regular media. We found that EMs express more Iba-1 compared to BMDMs and ANS4, but we were surprised to see that this marker was expressed neural progenitor cells, albeit at low levels (Supp. Fig. 6). Other studies have also noted that Iba-1 is not an exclusive marker of microglia as well (Szabo and Gulya, 2013; Bennett et al., 2016). However, comparisons of Iba-1 expression between EMs/ANS4, EMs, BMDMs/ANS4, BMDMs, and ANS4 cultures after 3 days did show higher expression levels of Iba-1 in EMs/ANS4 and BMDMs/ANS4 cocultures compared to EMs, BMDMs, and ANS4 cultured alone (Supp. Fig. 8). At day 20, we observed a higher density of Iba-1 positive cells per field of view in EMs/ANS4 and BMDMs/ANS4 co-cultures as compared to EMs, BMDMs, and ANS4 (Supp. Fig. 9). We have also observed a great expansion in the density of EMs and BMDMs between day 3 and day 20 when co-cultured with ANS4, while few cells where present in EMs or BMDMs cultured alone. To further determine the effects of NSPCs on the growth rate of EMs and BMDMs, we measured the cell density at different time points (1, 3, 6, 10, 13, 15, and 20 days). Quantification of the total numbers of cells per field of view showed that, starting at day 3 in culture, the numbers of EMs and BMDMs were significantly higher in co-culture conditions as compared to EMs and BMDMs cultured alone (Fig. 9W, Fig. 10O, and Supp. Fig. 11). This suggests that NSPCs have a trophic effect on macrophages that enhance macrophage proliferation and survival. This result is consistent with previous reports that neural stem cells produce wide array of secreted immune-modulatory and trophic molecules capable of promoting immune cell proliferation and survival (Kokaia et al., 2012).
The crosstalk between Macrophages and NSPCs is reciprocal
To determine if macrophages can also influence the differentiation of neural cells, we analyzed EM/ or BMDM/NSPC co-cultures for expression of the neural stem marker Nestin. Nestin is CNS-expressed intermediate filament protein that is down regulated upon differentiation of NSPCs and coincident with the increase in other intermediate filament proteins such as neurofilament in neurons and GFAP in astrocytes (Michalczyk and Ziman, 2005). First, we compared Nestin expression levels between EMs, BMDMs, and ANS4 cultured alone in their regular media without the influence of neural cues. Interestingly, we found that EMs, but not BMDMs are strongly positive for Nestin (Supp. Fig. 7). This result is consistent with data showing that yolk-sac-derived microglia in the retina, but not the circulating monocytes express Nestin (Ahmad, Tang and Pham, 2000; Wohl et al., 2011). Next, we examined Nestin expression in EMs/ANS4, EMs, BMDMs/ANS4, BMDMs, and ANS4 cultures under the influence of neural environment. After 3 days in culture, there were no differences in Nestin expression between EMs/ANS4, BMDMs/ANS4, and ANS4 cultured alone (Supp. Fig. 8), but after 20 days in culture, we observed morphological differences in the EMs/ANS4 and BMDMs/ANS4 co-cultures compared to ANS4 cultured alone. ANS4 cells continued to expand in culture as they do normally forming a near confluent monolayer of individual cells whereas cultures containing ANS4 cells with either EMs or BMDMs showed networks of cells with extended processes reminiscent of differentiated neurons (Supp. Fig. 9). To further confirm the differentiation of NSPCs into neurons, we examined ANS4, EMs/ANS4 and BMDMs/ANS4 cultures for their expression of the neuron specific beta class III tubulin (TUJ1), which is highly expressed in post-mitotic, differentiated neurons, and early neuronal precursors. At day 20, we found that ANS4 cells expressed higher levels of TUJ1 when co-cultured with EMs or BMDMs than ANS4 cultured alone (Supp. Fig. 10), indicating that co-culture of either EMs or BMDMs with ANS4 enhanced neuronal differentiation. Recent evidence points to an important role for microglia in regulating neurogenesis, neuronal differentiation, and neuronal circuit development (Butovsky et al., 2006; Miyamoto et al., 2016; Choi et al., 2017); however, the mechanisms by which EMs and BMDMs induced neuronal differentiation in our coculture system are unclear. Future studies are aimed at determining whether neural differentiation or neural network formation is promoted by specific cues made by microglia. Since it has been reported previously that microglia secret factors and cytokines that enhance neurogenesis in the subventricular zone (SVZ) during brain development, a comprehensive analysis of signaling molecules and cytokines from EMs will be necessary to fully understand the mechanisms underlying the effects of EMs on NSPCs proliferation and differentiation. Taken together, these studies show that not only are EMs influenced by neural cells, but the EMs may also supply specific cues that promoting CNS maturation during development.
CONCLUSIONS
Despite several studies reporting that yolk-sac-derived EMs play important roles in the development and maintenance of many tissues, including the vascular system, the exact cellular and molecular mechanisms, which confer these properties remain largely undefined. This is due to both a lack of suitable in vitro models of EMs and the fact that established adult macrophage lines are phenotypically and functionally distinct from EMs (Pucci et al., 2009; Wynn, Chawla and J. W. Pollard, 2013; Pinto et al., 2014; DeFalco et al., 2014). To overcome these issues, we have isolated and expanded EMs from E9.5 Csf1r-EGFP+/tg mice, characterized their molecular phenotype and assessed their angiogenic and differentiation potential as compared to adult BMDMs. The isolated EM population retains essential phenotypic and functional properties expected of yolksac-derived macrophages. The capability of mouse EMs to differentiate into bone fide microglia provide an approach to experimentally replace microglia after injury in mouse models of stroke, cerebral ischemia or neurological disorders. Several recent papers have shown that microglia can be generated from human iPS cells (Buchrieser, James and Moore, 2017; Douvaras et al., 2017; Takata et al., 2017). Interestingly, these microglia differentiate in a myb-independent fashion, similar to yolk sac-derived macrophages in the mouse. Purified cultures of mouse and human microglia will provide experimental approaches that can bridge between pre-clinical and translational models. Moreover, in vitro systems, such as the one described here will also help to improve our understanding of the molecular mechanisms underlying different roles of EMs during embryogenesis.
Supplementary Material
S1. Differential expression of pro-angiogenic and pro-inflammatory macrophage markers by EMs A. Fluorescence ICC images of BMDMs and EMs stained with CD11b (red) and DAPI (blue). B. Fluorescence ICC images of BMDMs and EMs stained with CCR2 (red) and DAPI (blue). C. Fluorescence ICC images of BMDMs and EMs stained with iNOS (red) and DAPI (blue). D. Fluorescence ICC images of BMDMs and EMs stained with Arg1 (red) and DAPI (blue). E. Fluorescence ICC images of BMDMs and EMs stained with C–3cr1 (red) and DAPI (blue). F. Fluorescence ICC images of BMDMs and EMs stained with Mrc1 (red) and DAPI (blue). G-I. Quantification of the CD11b, CCR2, and iNOS signals demonstrating significantly less EMs express CD11b, CCR2 and iNOS respectively as compared to BMDMs. J-L. Quantification of the Arg1, Cx3cr1, Mrc1 signals demonstrating significantly more EMs express Arg1, C–3cr1 and Mrc1 respectively as compared to BMDMs. Error bars indicate SE of the mean. N = 3. Scale bars 50 μm.
S7. Expression of the neural stem cell marker Nestin by EMs and BMDMs, and ANS4 in their regular medium. (A-C) Fluorescence ICC images of EMs, BMDMs, and ANS4 stained with Nestin (red). (D-F) Confocal images of Csf1r-EGFP transgene expression in EMs, BMDMs, and ANS4 (green) (G-I) EMs, BMDMs, and ANS4 stained with DAPI (blue). (J-L) Merged images EMs, BMDMs, and ANS4 Nestin antibody labeling, Csf1rEGFP transgene and DAPI staining.
S8. Expression of Iba-1 and Nestin by EMs and BMDMs, and ANS4 under the influence of neural environment at day 3. (A-E) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Iba-1 (yellow). (F-J) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Nestin (red). (K-O) Confocal images of Csf1r-EGFP transgene expression in EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 (green) (P-T) EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with DAPI (blue). (U-Y) Merged images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 Iba-1 and Nestin antibody labeling, Csf1r-EGFP transgene and DAPI staining.
S9. Expression of Iba-1 and Nestin by EMs and BMDMs, and ANS4 under the influence of neural environment at day 20. (A-E) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Iba-1 (yellow). (F-J) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Nestin (red). (K-O) Confocal images of Csf1r-EGFP transgene expression in EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 (green) (P-T) EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with DAPI (blue). (U-Y) Merged images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 Iba-1 and Nestin antibody labeling, Csf1r-EGFP transgene and DAPI staining.
S10. After 20 days, more ANS4 cells expressed the neuron specific beta class III tubulin (TUJ1) in the presence of macrophages. (A-C) Fluorescence ICC images of EMs + ANS4, BMDMs + ANS4, and ANS4 stained with TUJ1 (cherry). (D-F) Fluorescence ICC images of EMs + ANS4, BMDMs + ANS4, and ANS4 stained with Iba-1 (yellow). (G-I) Confocal images of Csf1r-EGFP transgene expression in EMs + ANS4, BMDMs + ANS4, and ANS4 (green) (J-L) EMs + ANS4, BMDMs + ANS4, and ANS4 stained with DAPI (blue). (M-O) Merged images of EMs + ANS4, BMDMs + ANS4, and ANS4 TUJ1 and Iba-1 antibody labeling, Csf1r-EGFP transgene and DAPI staining.
S11. Density measurements of EMs/BMDMs (Csf1r-EGFP+ cells) in EMs+ANS4, EMs, BMDMs+ANS4 and BMDMs cultures at day 1, 3, 6, 10, 13, 15, and 20. The data show starting at day 3 a higher density of EMs or BMDMs identified in groups that contained neural stem cells indicating that neural stem cells may support the survival or proliferation of EMs and BMDMs.
Table 1: Primary antibodies
Table 2: Secondary antibodies
S2. EMs and BMDMs showed equivalent levels of TNF-alpha immunostaining. Fluorescence ICC images of BMDMs stained with TNF-alpha (A), DAPI (B), and merge image of TNF-alpha and DAPI (C). Fluorescence ICC images of EMs stained with TNFalpha (D), DAPI (E), and merge image of TNF-alpha and DAPI (F). Scale bars 50 μm.
S3. EMs-HUVECs direct contact is necessary to promote cord formation. After 72 hours, the cord strustures formed by HUVECs fresh EGM complete medium (A), HUVECs incubated with EM conditioned medium (B), and HUVECs incubated with BMDM conditioned medium (C) imunnostained with PECAM-1 and imaged with confocal microscopy. Scale bars 500 μm.
S4. EMs did not differentiate into ramified microglia morphologies without the influence of neural environment at day 22. (A-C) Confocal images of EMs, and (C-F) confocal images of BMDMs displayed rounded and elongated cell forms, but no ramified microglia morphologies were observed in EMs and BMDMs cultures.
S5. Tmem119 is specifically expressed by EMs under neural environment at day 20. Comparative transcript expression analysis of Tmem119 in EMs, EMs+ANS4, BMDMs, BMDMs+ANS4, and ANS4 revealed Tmem119 was not detected in EMs or BMDMs cultured alone or with ANS4 at day 1. At day 20, both EMs and EMs+ANS4 expressed Tmem119, but EMs+ANS4 significantly expressed more Tmem119 compared to EMs cultured alone, while Tmem119 mRNA was not detected in BMDMs, BMDMs+ANS4, and ANS4. Error bars indicate SE of the mean. N = 3. ND, mRNA not detected.
S6. Expression of the microglia marker Iba-1 by EMs and BMDMs in non-neural environment. (A-C) Fluorescence ICC images of EMs, BMDMs, and ANS4 stained with Iba-1 (red). (D-F) Confocal images of Csf1r-EGFP transgene expression in EMs, BMDMs, and ANS4 (green) (G-I) EMs, BMDMs, and ANS4 stained with DAPI (blue). (JL) Merged images EMs, BMDMs, and ANS4 Iba-1 antibody labeling, Csf1r-EGFP transgene and DAPI staining.
ACKNOWLEDGEMENTS
The authors want to acknowledge the support of the Optical Imaging and Vital Microscopy Core at Baylor College of Medicine, and the support of the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574). We are grateful for support from the NIH (R01HL128064 and R01 EB016629 to MED) and the Cancer Prevention Research Institute of Texas (CPRIT MIRA award RP120713-P3 to MED).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmad I, Tang L and Pham H (2000) ‘Identification of neural progenitors in the adult mammalian eye.’, Biochemical and biophysical research communications, 270(2), pp. 517–521. doi: 10.1006/bbrc.2000.2473. [DOI] [PubMed] [Google Scholar]
- Ajami B, Bennett JL, Krieger C, Tetzlaff W and Rossi FMV (2007) ‘Local self-renewal can sustain CNS microglia maintenance and function throughout adult life.’, Nature neuroscience, 10(12), pp. 1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi Y, Lassoued W, Grama K, Nath SK, Zhu J, Oueslati R, Feldman M, Lee WMF and Roysam B (2011) ‘Cell-based quantification of molecular biomarkers in histopathology specimens’, Histopathology. Blackwell Publishing Ltd, 59(1), pp. 40–54. doi: 10.1111/j.1365-2559.2011.03878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold T and Betsholtz C (2013) ‘Correction: The importance of microglia in the development of the vasculature in the central nervous system.’, Vascular cell. BioMed Central, 5(1), p. 12. doi: 10.1186/2045-824X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG and Barres BA (2016) ‘New tools for studying microglia in the mouse and human CNS.’, Proceedings of the National Academy of Sciences of the United States of America. National Acad Sciences, 113(12), pp. E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Jalil A, Klaine M, Jung S, Cumano A and Godin I (2005) ‘Three pathways to mature macrophages in the early mouse yolk sac.’, Blood. American Society of Hematology, 106(9), pp. 3004–3011. doi: 10.1182/blood-2005-02-0461. [DOI] [PubMed] [Google Scholar]
- Buchrieser J, James W and Moore MD (2017) ‘Human Induced Pluripotent Stem Cell-Derived Macrophages Share Ontogeny with MYB-Independent Tissue-Resident Macrophages.’, Stem cell reports, 8(2), pp. 334–345. doi: 10.1016/j.stemcr.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Ziv Y, Schwartz A, Landa G, Talpalar AE, Pluchino S, Martino G and Schwartz M (2006) ‘Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells.’, Molecular and cellular neurosciences, 31(1), pp. 149–160. doi: 10.1016/j.mcn.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Choi JY, Kim JY, Kim JY, Park J, Lee WT and Lee JE (2017) ‘M2 Phenotype Microglia-derived Cytokine Stimulates Proliferation and Neuronal Differentiation of Endogenous Stem Cells in Ischemic Brain.’, Experimental neurobiology, 26(1), pp. 33–41. doi: 10.5607/en.2017.26.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T, Bhattacharya I, Williams AV, Sams DM and Capel B (2014) ‘Yolksac-derived macrophages regulate fetal testis vascularization and morphogenesis.’, Proceedings of the National Academy of Sciences of the United States of America. National Acad Sciences, 111(23), pp. E2384–93. doi: 10.1073/pnas.1400057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, Terrenoire C, Zhang B, Gandy S, Schadt E, Freytes DO, Noggle S and Fossati V (2017) ‘Directed Differentiation of Human Pluripotent Stem Cells to Microglia.’, Stem cell reports, 8(6), pp. 1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M, Steinfort DP, Smallwood D, Hew M, Chen W, Ernst M, Irving LB, Anderson GP and Hibbs ML (2016) ‘CD11b immunophenotyping identifies inflammatory profiles in the mouse and human lungs.’, Mucosal Immunology. Nature Publishing Group, 9(2), pp. 550–563. doi: 10.1038/mi.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder C, Schilling T, Heinemann U, Haas D, Hailer N and Nitsch R (1999) ‘Morphological, immunophenotypical and electrophysiological properties of resting microglia in vitro.’, The European journal of neuroscience, 11(12), pp. 4251–4261. [DOI] [PubMed] [Google Scholar]
- Eisenman ST, Gibbons SJ, Verhulst P-J, Cipriani G, Saur D and Farrugia G (2017) ‘Tumor necrosis factor alpha derived from classically activated “M1” macrophages reduces interstitial cell of Cajal numbers.’, Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society, 29(4), p. e12984. doi: 10.1111/nmo.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, Schilling JD, Schwendener R, Sergin I, Razani B, Forsberg EC, Yokoyama WM, Unanue ER, Colonna M, Randolph GJ and Mann DL (2014) ‘Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation.’, Immunity, 40(1), pp. 91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW and Ruhrberg C (2010) ‘Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction.’, Blood. American Society of Hematology, 116(5), pp. 829–840. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Tanaka J, Toku K, Tateishi N, Suzuki Y, Matsuda S, Sakanaka M and Maeda N (1996) ‘Effects of GM-CSF and ordinary supplements on the ramification of microglia in culture: a morphometrical study.’, Glia, 18(4), pp. 269–281. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M and Ley K (2010) ‘Development of monocytes, macrophages, and dendritic cells.’, Science (New York, N.Y.). American Association for the Advancement of Science, 327(5966), pp. 656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM and Merad M (2010) ‘Fate mapping analysis reveals that adult microglia derive from primitive macrophages.’, Science (New York, N.Y.). American Association for the Advancement of Science, 330(6005), pp. 841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F and Rodewald H-R (2015) ‘Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors.’, Nature, 518(7540), pp. 547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, Garc a-Sastre A, Stanley ER, Ginhoux F, Frenette PS and Merad M (2013) ‘Tissue-Resident Macrophages Self-Maintain Locally throughout Adult Life with Minimal Contribution from Circulating Monocytes’, Immunity, 38(4), pp. 792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Thisse B and Thisse C (2001) ‘Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process.’, Developmental biology. Academic Press, 238(2), pp. 274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- Ho VWH and Sly LM (2009) ‘Derivation and characterization of murine alternatively activated (M2) macrophages’, Methods in molecular biology (Clifton, N.J.). Totowa, NJ: Humana Press, 531(Chapter 12), pp. 173–185. doi: 10.1007/978-1-59745-396-7_12. [DOI] [PubMed] [Google Scholar]
- Hsu C-W, Poché RA, Saik JE, Ali S, Wang S, Yosef N, Calderon GA, Scott L, Vadakkan TJ, Larina IV, West JL and Dickinson ME (2015) ‘Improved Angiogenesis in Response to Localized Delivery of Macrophage-Recruiting Molecules.’, PloS one. Edited by E. Engel. Public Library of Science, 10(7), p. e0131643. doi: 10.1371/journal.pone.0131643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten N, Verbruggen S, Gijbels MJ, Post MJ, De Winther MPJ and Donners MMPC (2014) ‘Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo.’, Angiogenesis. Springer Netherlands, 17(1), pp. 109–118. doi: 10.1007/s10456-013-9381-6. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Kawamura N, Kumagai J, Fukuda J and Tanaka T (2007) ‘Tumor necrosis factor regulation of apoptosis in mouse preimplantation embryos and its antagonism by transforming growth factor alpha/phosphatidylionsitol 3-kinase signaling system.’, Biology of reproduction, 76(4), pp. 611–618. doi: 10.1095/biolreprod.106.058008. [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Hölscher C, Müller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, Rosenbauer F and Prinz M (2013) ‘Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways’, Nature neuroscience. Nature Publishing Group, 16(3), pp. 273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Kokaia Z, Martino G, Schwartz M and Lindvall O (2012) ‘Cross-talk between neural stem cells and immune cells: the key to better brain repair?’, Nature neuroscience, 15(8), pp. 1078–1087. doi: 10.1038/nn.3163. [DOI] [PubMed] [Google Scholar]
- Kroeger K, Collins M and Ugozzoli L (2009) ‘The preparation of primary hematopoietic cell cultures from murine bone marrow for electroporation.’, Journal of visualized experiments : JoVE, (23), pp. e1026–e1026. doi: 10.3791/1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larina IV, Shen W, Kelly OG, Hadjantonakis A-K, Baron MH and Dickinson ME (2009) ‘A membrane associated mCherry fluorescent reporter line for studying vascular remodeling and cardiac function during murine embryonic development.’, Anatomical record (Hoboken, N.J. : 2007). Wiley Subscription Services, Inc., A Wiley Company, 292(3), pp. 333–341. doi: 10.1002/ar.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitinger B and Hohenester E (2007) ‘Mammalian collagen receptors.’, Matrix biology : journal of the International Society for Matrix Biology, 26(3), pp. 146–155. doi: 10.1016/j.matbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lichanska AM and Hume DA (2000) ‘Origins and functions of phagocytes in the embryo.’, Experimental hematology, 28(6), pp. 601–611. [DOI] [PubMed] [Google Scholar]
- Lichanska AM, Browne CM, Henkel GW, Murphy KM, Ostrowski MC, McKercher SR, Maki RA and Hume DA (1999) ‘Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.1.’, Blood, 94(1), pp. 127–138. [PubMed] [Google Scholar]
- Lin EY and Pollard JW (2007) ‘Tumor-associated macrophages press the angiogenic switch in breast cancer.’, Cancer research. American Association for Cancer Research, 67(11), pp. 5064–5066. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- Lin EY, Li J-F, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, Qian H, Xue X-N and Pollard JW (2006) ‘Macrophages regulate the angiogenic switch in a mouse model of breast cancer.’, Cancer research. American Association for Cancer Research, 66(23), pp. 11238–11246. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- Liu J, Xue Y, Dong D, Xiao C, Lin C, Wang H, Song F, Fu T, Wang Z, Chen J, Pan H, Li Y, Cai D and Li Z (2017) ‘CCR2? and CCR2+ corneal macrophages exhibit distinct characteristics and balance inflammatory responses after epithelial abrasion’, Mucosal Immunology. Nature Publishing Group, 46, p. 845. doi: 10.1038/mi.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen DH and Bugge TH (2013) ‘Imaging collagen degradation in vivo highlights a key role for M2-polarized macrophages in extracellular matrix degradation.’, Oncoimmunology. Taylor & Francis, 2(12), p. e27127. doi: 10.4161/onci.27127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen DH, Leonard D, Masedunskas A, Moyer A, Jürgensen HJ, Peters DE, Amornphimoltham P, Selvaraj A, Yamada SS, Brenner DA, Burgdorf S, Engelholm LH, Behrendt N, Holmbeck K, Weigert R and Bugge TH (2013) ‘M2-like macrophages are responsible for collagen degradation through a mannose receptor-mediated pathway.’, The Journal of cell biology. Rockefeller University Press, 202(6), pp. 951–966. doi: 10.1083/jcb.201301081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A and Locati M (2004) ‘The chemokine system in diverse forms of macrophage activation and polarization.’, Trends in immunology, 25(12), pp. 677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Marim FM, Silveira TN, Lima DS and Zamboni DS (2010) ‘A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells.’, PloS one. Edited by P. T. Bozza. Public Library of Science, 5(12), p. e15263. doi: 10.1371/journal.pone.0015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, Jacome-Galarza CE, Handler K, Klughammer J, Kobayashi Y, Gomez-Perdiguero E, Schultze JL, Beyer M, Bock C and Geissmann F (2016) ‘Specification of tissueresident macrophages during organogenesis’, Science (New York, N.Y.). American Association for the Advancement of Science, 353(6304), pp. aaf4238–aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, Elinav E, Sieweke MH, Schwartz M and Amit I (2016) ‘Microglia development follows a stepwise program to regulate brain homeostasis.’, Science (New York, N.Y.). American Association for the Advancement of Science, 353(6301), pp. aad8670–aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- Mazzieri R, Pucci F, Moi D, Zonari E, Ranghetti A, Berti A, Politi LS, Gentner B, Brown JL, Naldini L and De Palma M (2011) ‘Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells.’, Cancer cell, 19(4), pp. 512–526. doi: 10.1016/j.ccr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- McGrath KE, Frame JM and Palis J (2015) ‘Early hematopoiesis and macrophage development.’, Seminars in immunology, 27(6), pp. 379–387. doi: 10.1016/j.smim.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczyk K and Ziman M (2005) ‘Nestin structure and predicted function in cellular cytoskeletal organisation.’, Histology and histopathology, 20(2), pp. 665–671. doi: 10.14670/HH-20.665. [DOI] [PubMed] [Google Scholar]
- Miyamoto A, Wake H, Ishikawa AW, Eto K, Shibata K, Murakoshi H, Koizumi S, Moorhouse AJ, Yoshimura Y and Nabekura J (2016) ‘Microglia contact induces synapse formation in developing somatosensory cortex’, Nature Communications. Nature Publishing Group, 7, p. 12540. doi: 10.1038/ncomms12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM and Cardona AE (2012) ‘The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood.’, Journal of immunology (Baltimore, Md. : 1950). American Association of Immunologists, 188(1), pp. 29–36. doi: 10.4049/jimmunol.1100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH and Zon LI (2008) ‘Hematopoiesis: an evolving paradigm for stem cell biology.’, Cell, 132(4), pp. 631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov DA (2008) ‘Macrophages in the embryo and beyond: much more than just giant phagocytes.’, Genesis (New York, N.Y. : 2000). Wiley Subscription Services, Inc., A Wiley Company, 46(9), pp. 447–462. doi: 10.1002/dvg.20417. [DOI] [PubMed] [Google Scholar]
- Owen JL and Mohamadzadeh M (2013) ‘Macrophages and chemokines as mediators of angiogenesis.’, Frontiers in physiology. Frontiers, 4, p. 159. doi: 10.3389/fphys.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AR, Godwin JW, Chandran A, Hersey L, Ilinykh A, Debuque R, Wang L and Rosenthal NA (2014) ‘Age-related changes in tissue macrophages precede cardiac functional impairment.’, Aging, 6(5), pp. 399–413. doi: 10.18632/aging.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poché RA, Hsu C-W, McElwee ML, Burns AR and Dickinson ME (2015) ‘Macrophages engulf endothelial cell membrane particles preceding pupillary membrane capillary regression.’, Developmental biology, 403(1), pp. 30–42. doi: 10.1016/j.ydbio.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SM (2013) ‘In vitro expansion of fetal neural progenitors as adherent cell lines’, Methods in molecular biology (Clifton, N.J.). Totowa, NJ: Humana Press, 1059(Chapter 2), pp. 13–24. doi: 10.1007/978-1-62703-574-3_2. [DOI] [PubMed] [Google Scholar]
- Pucci F, Venneri MA, Biziato D, Nonis A, Moi D, Sica A, Di Serio C, Naldini L and De Palma M (2009) ‘A distinguishing gene signature shared by tumor-infiltrating Tie2-expressing monocytes, blood “resident” monocytes, and embryonic macrophages suggests common functions and developmental relationships.’, Blood. American Society of Hematology, 114(4), pp. 901–914. doi: 10.1182/blood-2009-01-200931. [DOI] [PubMed] [Google Scholar]
- Rao S, Lobov IB, Vallance JE, Tsujikawa K, Shiojima I, Akunuru S, Walsh K, Benjamin LE and Lang RA (2007) ‘Obligatory participation of macrophages in an angiopoietin 2-mediated cell death switch.’, Development (Cambridge, England). The Company of Biologists Ltd, 134(24), pp. 4449–4458. doi: 10.1242/dev.012187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rőszer T (2015) ‘Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms.’, Mediators of inflammation. Hindawi Publishing Corporation, 2015(4), pp. 816460–16. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymo SF, Gerhardt H, Wolfhagen Sand F, Lang R, Uv A and Betsholtz C (2011) ‘A Two-Way Communication between Microglial Cells and Angiogenic Sprouts Regulates Angiogenesis in Aortic Ring Cultures’, PloS one. Edited by M. O. Karl. Public Library of Science, 6(1), p. e15846. doi: 10.1371/journal.pone.0015846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, Ostrowski MC, Himes SR and Hume DA (2003) ‘A macrophage colonystimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse.’, Blood. American Society of Hematology, 101(3), pp. 1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Schmieder A, Michel J, Schönhaar K, Goerdt S and Schledzewski K (2012) ‘Differentiation and gene expression profile of tumor-associated macrophages.’, Seminars in cancer biology, 22(4), pp. 289–297. doi: 10.1016/j.semcancer.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW, Frampton J, Liu KJ and Geissmann F (2012) ‘A lineage of myeloid cells independent of Myb and hematopoietic stem cells.’, Science (New York, N.Y.). American Association for the Advancement of Science, 336(6077), pp. 86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Sheets KG, Jun B, Zhou Y, Zhu M, Petasis NA, Gordon WC and Bazan NG (2013) ‘Microglial ramification and redistribution concomitant with the attenuation of choroidal neovascularization by neuroprotectin D1.’, Molecular vision. Emory University, 19, pp. 1747–1759. [PMC free article] [PubMed] [Google Scholar]
- Szabo M and Gulya K (2013) ‘Development of the microglial phenotype in culture.’, Neuroscience, 241, pp. 280–295. doi: 10.1016/j.neuroscience.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Donovan MJ, Rogers RA and Ezekowitz RA (1998) ‘Distribution of murine mannose receptor expression from early embryogenesis through to adulthood.’, Cell and tissue research, 292(2), pp. 311–323. [DOI] [PubMed] [Google Scholar]
- Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, Chakarov S, See P, Low D, Low G, Garcia-Miralles M, Zeng R, Zhang J, Goh CC, Gul A, Hubert S, Lee B, Chen J, Low I, Shadan NB, Lum J, Wei TS, Mok E, Kawanishi S, Kitamura Y, Larbi A, Poidinger M, Renia L, Ng LG, Wolf Y, Jung S, Önder T, Newell E, Huber T, Ashihara E, Garel S, Pouladi MA and Ginhoux F (2017) ‘Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function.’, Immunity, 47(1), pp. 183–198.e6. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Tammela T, Zarkada G, Nurmi H, Jakobsson L, Heinolainen K, Tvorogov D, Zheng W, Franco CA, Murtomäki A, Aranda E, Miura N, Ylä-Herttuala S, Fruttiger M, Mäkinen T, Eichmann A, Pollard JW, Gerhardt H and Alitalo K (2011) ‘VEGFR-3 controls tip to stalk conversion at vessel fusion sites by reinforcing Notch signalling.’, Nature cell biology, 13(10), pp. 1202–1213. doi: 10.1038/ncb2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Narayanaswamy A, Tsai C-L and Roysam B (2011) ‘A broadly applicable 3-D neuron tracing method based on open-curve snake.’, Neuroinformatics. Springer-Verlag, 9(2–3), pp. 193–217. doi: 10.1007/s12021-011-9110-5. [DOI] [PubMed] [Google Scholar]
- Willenborg S, Lucas T, van Loo G, Knipper JA, Krieg T, Haase I, Brachvogel B, Hammerschmidt M, Nagy A, Ferrara N, Pasparakis M and Eming SA (2012) ‘CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair.’, Blood. American Society of Hematology, 120(3), pp. 613–625. doi: 10.1182/blood-2012-01-403386. [DOI] [PubMed] [Google Scholar]
- Wohl SG, Schmeer CW, Friese T, Witte OW and Isenmann S (2011) ‘In situ dividing and phagocytosing retinal microglia express nestin, vimentin, and NG2 in vivo.’, PloS one. Edited by R. Linden. Public Library of Science, 6(8), p. e22408. doi: 10.1371/journal.pone.0022408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wride MA and Sanders EJ (1995) ‘Potential roles for tumour necrosis factor? during embryonic development’, Anatomy and Embryology. Springer-Verlag, 191(1), pp. 1–10. doi: 10.1007/BF00215292. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A and Pollard JW (2013) ‘Macrophage biology in development, homeostasis and disease.’, Nature, 496(7446), pp. 445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Differential expression of pro-angiogenic and pro-inflammatory macrophage markers by EMs A. Fluorescence ICC images of BMDMs and EMs stained with CD11b (red) and DAPI (blue). B. Fluorescence ICC images of BMDMs and EMs stained with CCR2 (red) and DAPI (blue). C. Fluorescence ICC images of BMDMs and EMs stained with iNOS (red) and DAPI (blue). D. Fluorescence ICC images of BMDMs and EMs stained with Arg1 (red) and DAPI (blue). E. Fluorescence ICC images of BMDMs and EMs stained with C–3cr1 (red) and DAPI (blue). F. Fluorescence ICC images of BMDMs and EMs stained with Mrc1 (red) and DAPI (blue). G-I. Quantification of the CD11b, CCR2, and iNOS signals demonstrating significantly less EMs express CD11b, CCR2 and iNOS respectively as compared to BMDMs. J-L. Quantification of the Arg1, Cx3cr1, Mrc1 signals demonstrating significantly more EMs express Arg1, C–3cr1 and Mrc1 respectively as compared to BMDMs. Error bars indicate SE of the mean. N = 3. Scale bars 50 μm.
S7. Expression of the neural stem cell marker Nestin by EMs and BMDMs, and ANS4 in their regular medium. (A-C) Fluorescence ICC images of EMs, BMDMs, and ANS4 stained with Nestin (red). (D-F) Confocal images of Csf1r-EGFP transgene expression in EMs, BMDMs, and ANS4 (green) (G-I) EMs, BMDMs, and ANS4 stained with DAPI (blue). (J-L) Merged images EMs, BMDMs, and ANS4 Nestin antibody labeling, Csf1rEGFP transgene and DAPI staining.
S8. Expression of Iba-1 and Nestin by EMs and BMDMs, and ANS4 under the influence of neural environment at day 3. (A-E) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Iba-1 (yellow). (F-J) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Nestin (red). (K-O) Confocal images of Csf1r-EGFP transgene expression in EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 (green) (P-T) EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with DAPI (blue). (U-Y) Merged images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 Iba-1 and Nestin antibody labeling, Csf1r-EGFP transgene and DAPI staining.
S9. Expression of Iba-1 and Nestin by EMs and BMDMs, and ANS4 under the influence of neural environment at day 20. (A-E) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Iba-1 (yellow). (F-J) Fluorescence ICC images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with Nestin (red). (K-O) Confocal images of Csf1r-EGFP transgene expression in EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 (green) (P-T) EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 stained with DAPI (blue). (U-Y) Merged images of EMs + ANS4, EMs, BMDMs + ANS4, BMDMs, and ANS4 Iba-1 and Nestin antibody labeling, Csf1r-EGFP transgene and DAPI staining.
S10. After 20 days, more ANS4 cells expressed the neuron specific beta class III tubulin (TUJ1) in the presence of macrophages. (A-C) Fluorescence ICC images of EMs + ANS4, BMDMs + ANS4, and ANS4 stained with TUJ1 (cherry). (D-F) Fluorescence ICC images of EMs + ANS4, BMDMs + ANS4, and ANS4 stained with Iba-1 (yellow). (G-I) Confocal images of Csf1r-EGFP transgene expression in EMs + ANS4, BMDMs + ANS4, and ANS4 (green) (J-L) EMs + ANS4, BMDMs + ANS4, and ANS4 stained with DAPI (blue). (M-O) Merged images of EMs + ANS4, BMDMs + ANS4, and ANS4 TUJ1 and Iba-1 antibody labeling, Csf1r-EGFP transgene and DAPI staining.
S11. Density measurements of EMs/BMDMs (Csf1r-EGFP+ cells) in EMs+ANS4, EMs, BMDMs+ANS4 and BMDMs cultures at day 1, 3, 6, 10, 13, 15, and 20. The data show starting at day 3 a higher density of EMs or BMDMs identified in groups that contained neural stem cells indicating that neural stem cells may support the survival or proliferation of EMs and BMDMs.
Table 1: Primary antibodies
Table 2: Secondary antibodies
S2. EMs and BMDMs showed equivalent levels of TNF-alpha immunostaining. Fluorescence ICC images of BMDMs stained with TNF-alpha (A), DAPI (B), and merge image of TNF-alpha and DAPI (C). Fluorescence ICC images of EMs stained with TNFalpha (D), DAPI (E), and merge image of TNF-alpha and DAPI (F). Scale bars 50 μm.
S3. EMs-HUVECs direct contact is necessary to promote cord formation. After 72 hours, the cord strustures formed by HUVECs fresh EGM complete medium (A), HUVECs incubated with EM conditioned medium (B), and HUVECs incubated with BMDM conditioned medium (C) imunnostained with PECAM-1 and imaged with confocal microscopy. Scale bars 500 μm.
S4. EMs did not differentiate into ramified microglia morphologies without the influence of neural environment at day 22. (A-C) Confocal images of EMs, and (C-F) confocal images of BMDMs displayed rounded and elongated cell forms, but no ramified microglia morphologies were observed in EMs and BMDMs cultures.
S5. Tmem119 is specifically expressed by EMs under neural environment at day 20. Comparative transcript expression analysis of Tmem119 in EMs, EMs+ANS4, BMDMs, BMDMs+ANS4, and ANS4 revealed Tmem119 was not detected in EMs or BMDMs cultured alone or with ANS4 at day 1. At day 20, both EMs and EMs+ANS4 expressed Tmem119, but EMs+ANS4 significantly expressed more Tmem119 compared to EMs cultured alone, while Tmem119 mRNA was not detected in BMDMs, BMDMs+ANS4, and ANS4. Error bars indicate SE of the mean. N = 3. ND, mRNA not detected.
S6. Expression of the microglia marker Iba-1 by EMs and BMDMs in non-neural environment. (A-C) Fluorescence ICC images of EMs, BMDMs, and ANS4 stained with Iba-1 (red). (D-F) Confocal images of Csf1r-EGFP transgene expression in EMs, BMDMs, and ANS4 (green) (G-I) EMs, BMDMs, and ANS4 stained with DAPI (blue). (JL) Merged images EMs, BMDMs, and ANS4 Iba-1 antibody labeling, Csf1r-EGFP transgene and DAPI staining.