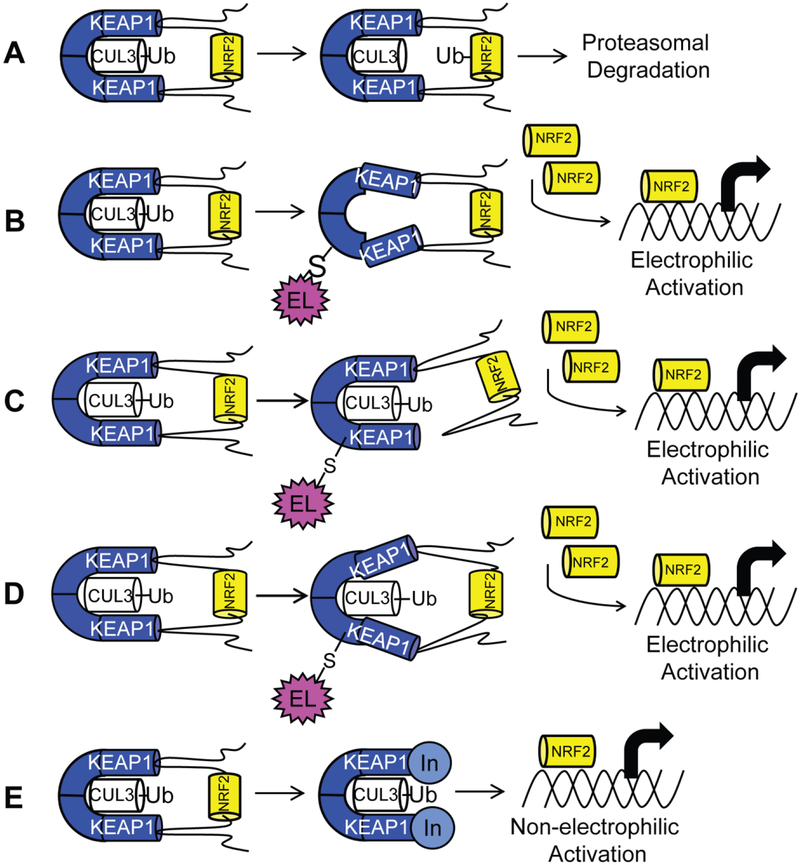
Figure 1:
(A) Transcription factor NRF2, when bound to substrate adaptor KEAP1, is ubiquitinated by E3 ligase CUL3, leading to proteosomal degradation. Two mechanisms explain activation of NRF2 by electrophiles. (B) In the CUL3 dissociation mechanism, cysteine modification by an electrophile causes dissociation of CUL3, leading to accumulation of NRF2. (C) In the hinge-and-latch mechanism, cysteine modification causes non-optimal placement of NRF2 for degradation. (D) In the conformational cycling model, cysteine modification locks KEAP1 and NRF2 in a conformation not ideal for ubiquitination, preventing KEAP1 from binding nascent NRF2, leading to accumulation of NRF2 (E) The non-electrophilic mechanism aims to create a non-covalent inhibitor of the protein-protein interaction between KEAP1 and NRF2 to activate NRF2.