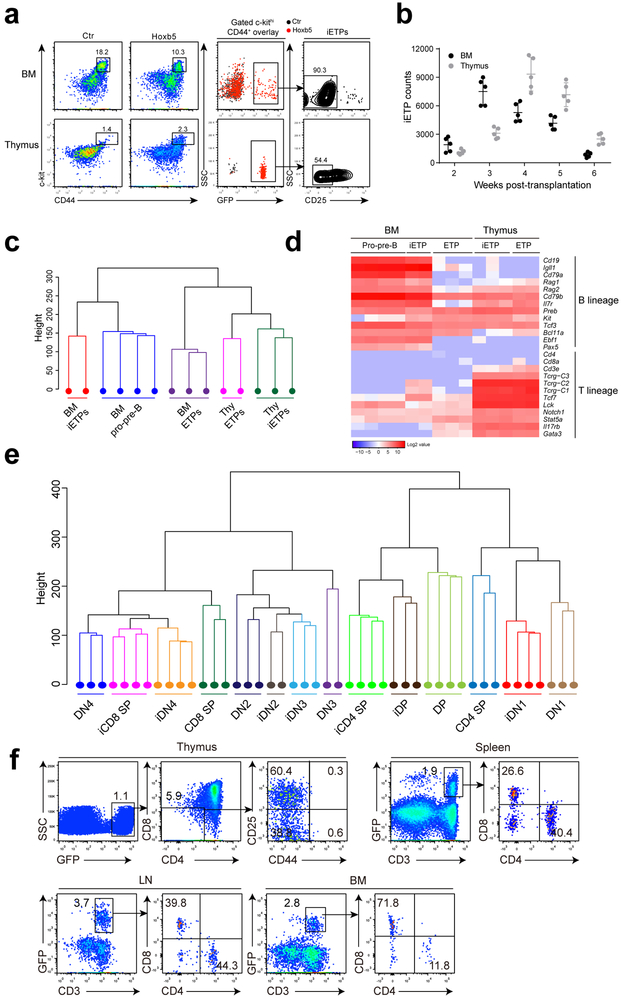
Figure 7. Hoxb5 directly converts B lymphocytes to early T cell progenitor-like cells in bone marrow.
(a) Flow cytometry analysis of iETPs in the BM and thymus of a representative retro-Hoxb5 recipient four weeks post-transplantation. Five million sorted retro-Hoxb5 pro-pre-B cells were transplanted into sublethally irradiated individual recipients. Recipients transplanted with pro-pre-B cells transduced with GFP control virus were used as controls. Representative plots from five mice of each groups were shown. (b) Absolute numbers of iETP cells in the BM and thymus of Hoxb5 recipients (n = 5 mice) at different time points after transplantation. Each symbol represents an individual mouse, and small horizontal lines indicate the mean (± s.d.). (c) Unsupervised hierarchical clustering of RNA-Seq data of iETPs, natural ETPs and pro-pre-B cells. For each RNA-Seq sample, one thousand iETPs or ETP (Lin−CD44+c-kithiCD25−) from retro-Hoxb5 mice or wild type control mice were sorted and analysed. (d) Heatmaps showed the expression pattern of selected genes related to T or B cells between iETPs, natural ETPs and pro-pre-B cells. Columns represent the indicated cell subsets in biological replicates. (e) Unsupervised hierarchical clustering of RNASeq data of iDN1, iDN2, iDN3, iDN4, iDP, iCD4-SP, iCD8-SP cells and related wild type counterparts. (f) Flow cytometry analysis of iT lymphocytes in thymus, spleen, LN and BM of a representative recipient 3 weeks after secondary transplantation of iT thymocytes. Data are representative of two independent experiments.