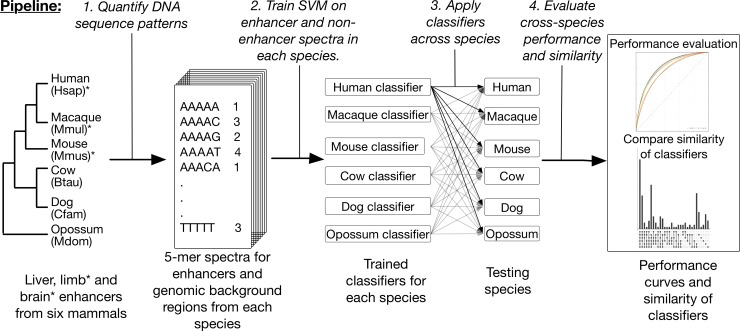
Fig 1. Overview of the framework for evaluating DNA patterns predictive of enhancer activity across diverse mammals.
Starting with liver, limb and brain enhancers and genomic background regions from six mammals, the first step of the pipeline quantified each of these genomic regions by their 5-mer spectrum—the frequency of occurrence of all possible length five DNA sequence patterns. Using the spectra as features, we trained a spectrum kernel support vector machine (SVM) to distinguish enhancers from non-enhancers in each species and evaluated their performance with ten-fold cross validation. Then, we applied classifiers trained on one species to predict enhancer activity in all other species. Finally, we evaluated the performance of cross-species prediction compared to within species prediction and quantified the similarity of different species’ classifiers by the sharing of TF motifs among the most predictive 5-mers. Limb and brain enhancer data were only available for human, macaque, and mouse.