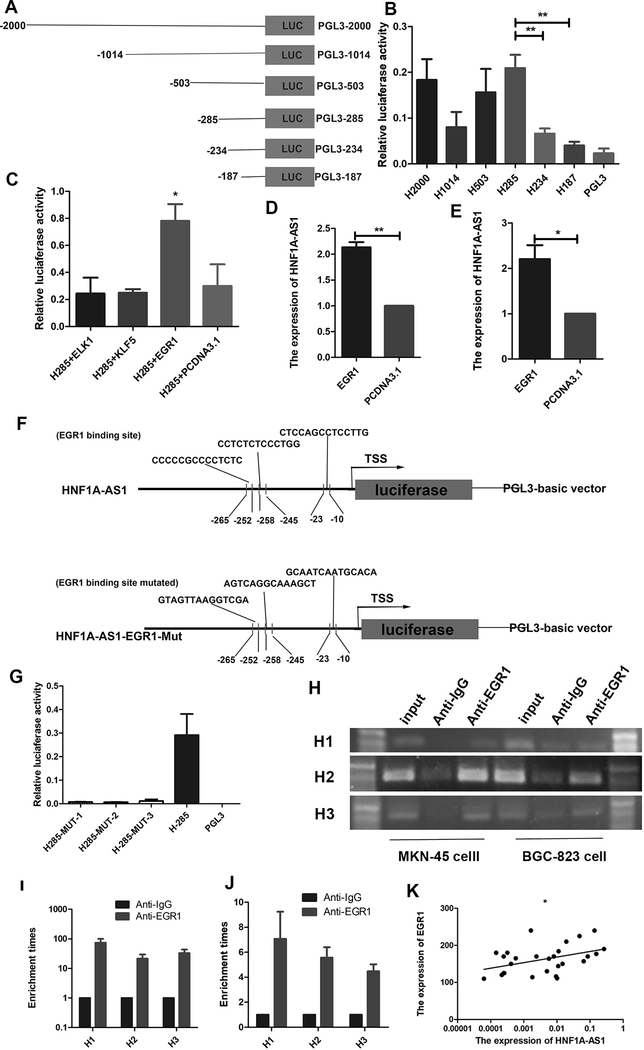
Figure 3. EGR1 activates HNF1A-AS1 transcription.
(A). Schematic diagram of the HNF1A-AS1 promoter fragments spanning from −2,000/ −1,014/ −503/ −285/ −234/ −187 to 0. This promoter fragments were cloned into the upstream of the firefly luciferase reporter gene in the pGL3-basic vector.
(B). Transcriptional activity analysis of the potential HNF1A-AS1 promoter fragments in 293T cells.
(C). Luciferase activity assay showed that EGR1 observably increased promoter activities of pGL3−285/0.
(D-E). RT-qPCR assay indicated EGR1 promoted the expression level of HNF1A-AS1 in MKN-45 cells (D) and BGC-823 cells (E).
(F). Schematic diagram of the luciferase reporter construct containing the human HNF1A-AS1 promoter and the mutant construct (H-Mut-1/2/3) containing the basal promoter in which the presumed HNF1A-AS1 binding site was mutated.
(G). Luciferase activity of the HNF1A-AS1 promoter was decreased when the three presumed HNF1A-AS1 binding site was mutated.
(H). ChIP-PCR assay showed that EGR1directly interacted with the EGR1 binding sites within HNF1A-AS1 promoter in MKN-45 cells and BGC-823 cells. A specific strong band of the expected size was detected in the input DNA. The fragment containing the EGR1 binding sites was detected. No band or very weak band was detected in the chromatin complex precipitated by antibody against IgG. H1, H2 and H3 respectively represent primer that covered the EGR1 binding sites.
(I-J). ChIP-qPCR analysis indicated higher fold enrichment of promoter amplicons of EGR1 in anti-EGR1 antibody group than that of IgG group in MKN-45 cells (I) and BGC-823 cells (J), indicating that EGR1 could directly bind to HNF1A-AS1 promoter. H1, H2 and H3 respectively represent primer that covered the EGR1 binding sites.
(K). A significant positive correlation was found between the protein levels of EGR1 and HNF1A-AS1 in GC tissues. Three independent experiments were performed, and data are presented as mean ± SD.
*P < 0.05, ** P < 0.01, *** P < 0.001