Figure 2. Reconstruction of a brain network.
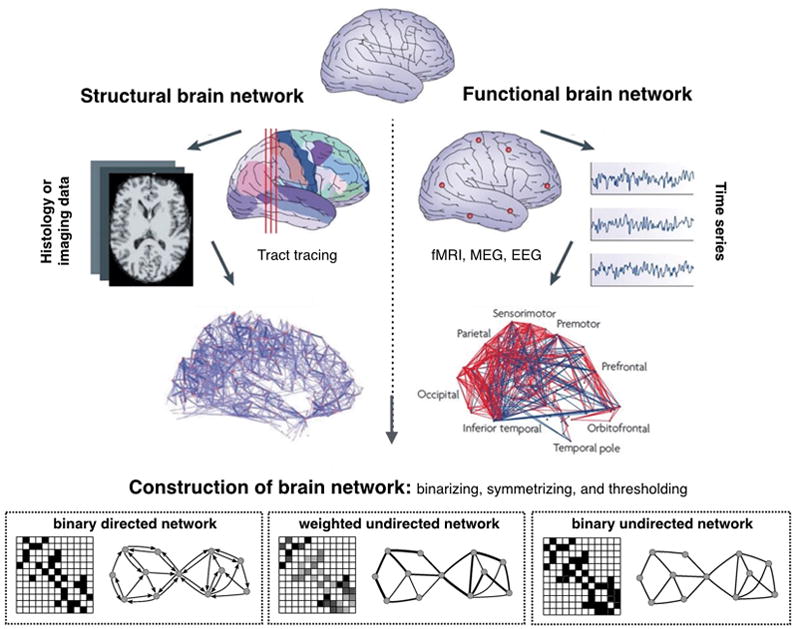
Step 1: Define the network nodes. These could be defined as electroencephalography sources or multi-electrode-array electrodes as well as anatomically defined regions of histological, MRI or diffusion tensor imaging data.
Step 2: Define the network edges. Estimate a continuous measure of association between nodes. This could be the spectral coherence or Granger causality measures between two magnetoencephalography sensors, or the connection probability between two regions of an individual diffusion tensor imaging data set, or the inter-regional correlations in cortical thickness or volume MRI measurements estimated in groups of subjects.
Step 3: Define the network structure. Generate an association matrix by compiling all pairwise associations between nodes and apply a threshold to each element of this matrix to produce a binary or undirected or directed network.
Step 4: Calculate the network properties (path length, clustering coefficient, modularity, etc.) of interest in this graphical model of a brain network.
