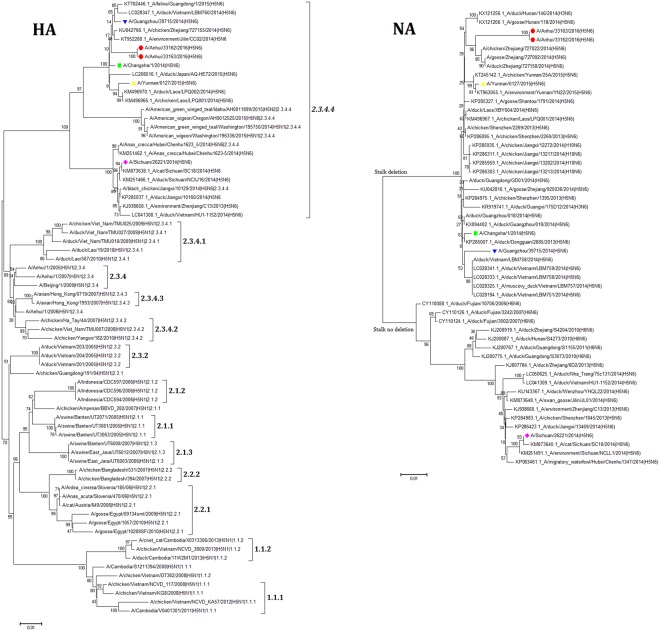
Figure 1.
Phylogenetic trees of avian influenza A viruses, based on the haemagglutinin (HA) and neuraminidase (NA) genes. Neighbour-joining trees were constructed using the Molecular Evolutionary Genetics Analysis software (MEGA 6.0). A dendrogram was obtained based on genetic distances. All reference sequences were downloaded from NCBI, GISAID or the National Influenza Centre of China. Bootstrap values were calculated for 1,000 replicates. A/Anhui/33162/2016(H5N6) and A/Anhui/33163/2016(H5N6) were indicated with red dots. A/Sichuan/26221/2014(H5N6), A/Yunnan/0127/2015(H5N6), A/Guangdong/39715/2014(H5N6) and A/Changsha/1/2014(H5N6) were indicated with a pink rhombus, a yellow positive trigonometric, a blue inverted triangle and a green square, respectively. Brackets denote H5 subtype virus clades. Scale bars indicate nucleotide substitutions per site.