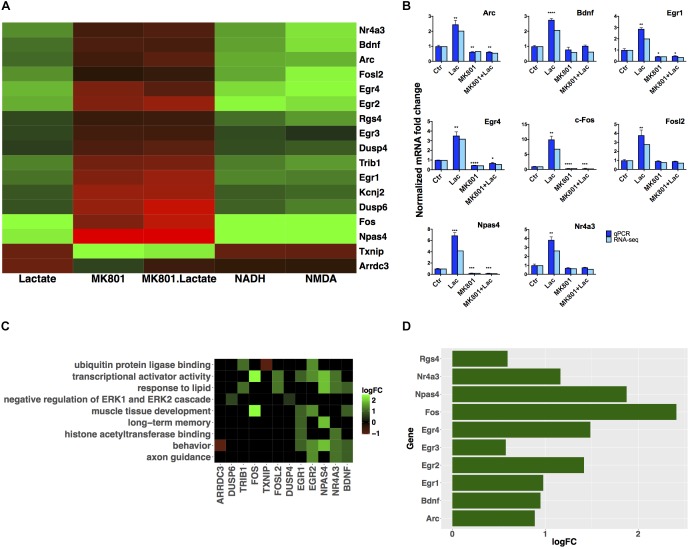
FIGURE 3.
Subset of genes modulated by L-lactate. (A) Fold change modulation of genes regulated by L-lactate in presence and absence of MK-801, and by NADH, NMDA, and MK-801 alone. (B) Validation of RNA-seq data with qRT-PCR, comparison of fold expression changes between RNA-seq and qRT-PCR data for genes is shown. Error bars correspond to standard error of the mean (SEM) values. T-test conducted for qRT-PCR analysis, between the different treatments and Ctr condition; significance (p < 0.05∗, p < 0.01∗∗, p < 0.001∗∗∗, p < 0.0001∗∗∗∗). (C) Genes-terms relationship heatmap emphasizing genes associated with multiple gene ontology terms of interest and showing their fold changes expression in response to L-lactate treatment at 1 h. Up-regulated genes are indicated in green and down-regulated genes in red. All terms displayed are over-represented with a false discovery rate (FDR) < 0.05. (D) Bar plot showing fold induction of a few selected known synaptic plasticity-related genes. The x-axis shows gain of expression log2(fold change) 1 h after L-lactate stimulation.