FIGURE 3.
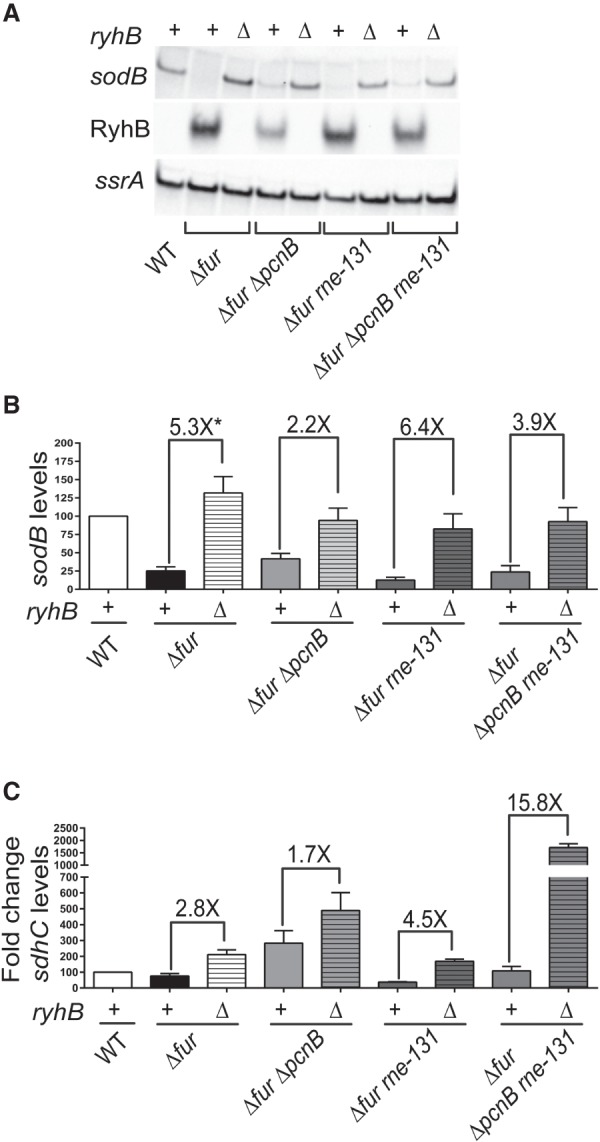
In the absence of poly(A) polymerase, RyhB does not efficiently regulate sodB and sdhCDAB target mRNAs. (A,B) Northern blot analysis was used to determine RyhB and sodB steady state in the wild-type and derived isogenic mutant strains, and (C) qRT-PCR analysis was used to determine sdhC levels. Wild-type parent (WT [fur+]; NRD1138) and its derived isogenic mutants (Δfur, DS024; Δfur ΔpcnB, DS025; Δfur rne-131, DS069; Δfur ΔpcnB rne-131, DS082; Δfur ΔryhB, NRD1546; Δfur ΔpcnB ΔryhB, NRD1547; Δfur rne-131 ΔryhB, NRD1550; Δfur ΔpcnB rne-131 ΔryhB, NRD1551) were diluted 200-fold in fresh MOPS EZ rich defined media supplemented with 0.4% glycerol and grown to late exponential phase (OD600 of 1.0), and samples for RNA extraction were collected. Representative northern blots are shown (A). (B,C) Graphs are presented that display the relative expression levels of sodB and sdhC mRNA in a wild-type strain and derived mutant strains. Briefly, the signal intensities for sodB and sdhC transcripts were first quantified from northern blots or qRT-PCRs. The signal intensity was then normalized to the ssrA transcript level, which served as the loading control, and subsequently the expression level relative to the fur+ (WT) strain NRD1138, which was set at 100%, was calculated. sdhC transcript fold changes relative to NRD1138 were calculated via the ΔΔCt method. Asterisk (*) in B indicates that the calculated fold change is >5.3× since sodB steady-state determination in DS024 (Δfur) strain was not accurate due to very low signal intensity. Data shown in B and C represent the mean (±SEM) of at least three independent experiments. Probes and primers used are listed in Supplemental Table S2.
