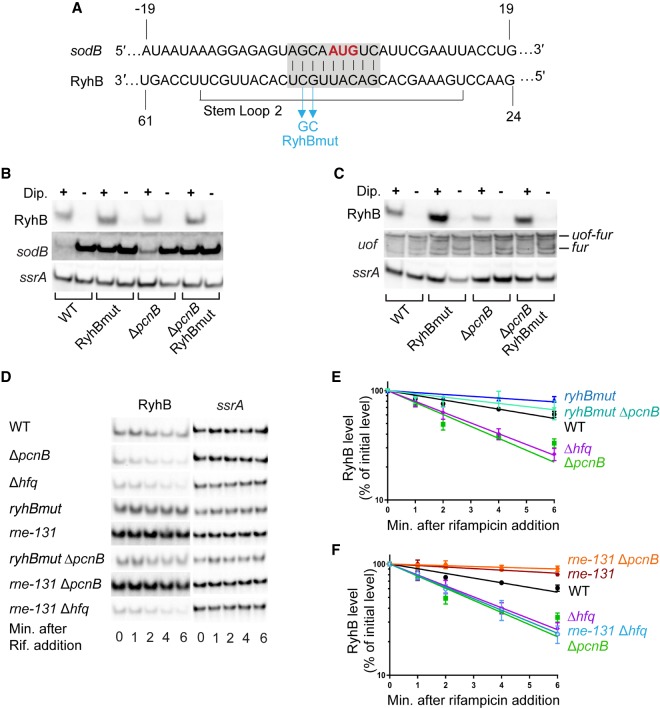
FIGURE 5.
The instability of RyhB in the absence of poly(A) polymerase is due to pairing with target RNAs. (A) Schematic showing complementarity (highlighted in gray) between RyhB and its target mRNA sodB, and the specific mutations (in cyan) introduced in RyhB to create a RyhB variant unable to pair with target mRNAs (RyhBmut). The start codon of sodB is highlighted in red. (B,C) Northern blot analysis to determine the transcript steady-state levels of RyhB targets sodB and uof in a wild-type strain (WT [fur+]; NRD1138) and its derived isogenic mutants (ΔpcnB, NRD1198; ryhBmut, LM11; ryhBmut ΔpcnB) under RyhB inducing and noninducing conditions. Overnight cultures of these strains grown in LB were diluted 200-fold in fresh LB media and grown to log phase and treated with dipyridyl (+) to chelate iron or mock-treated (−). After 15 min of treatment, RNA was extracted from each culture and prepared for northern blot analysis to determine corresponding levels of sodB, uof, RyhB, and ssrA (loading control). uof transcription is driven from two upstream promoters Puof and Pfur to generate the uof-fur and fur mRNAs, respectively, but RyhB specifically interacts with the fur mRNA (C). (D–F) Determination of RyhB intrinsic stability in a wild-type (WT [fur+]; NRD1138) and its derived isogenic mutants (ΔpcnB, NRD1198; Δhfq, DS021; ryhBmut, LM11; ryhBmut ΔpcnB, LM13; rne-131, DS102; rne-131 ΔpcnB, DS106; rne-131 Δhfq, DS130). Strains were subjected to RNA stability time-course experiments as described in the legend of Figure 4. Representative northern blots are shown in D. (E,F) RyhB decay curves were generated as described in Figure 4 and corresponding half-life measurements are listed in Table 1. Points and error bars in the curves represent the means and the standard errors (SEM) of at least three independent experiments.