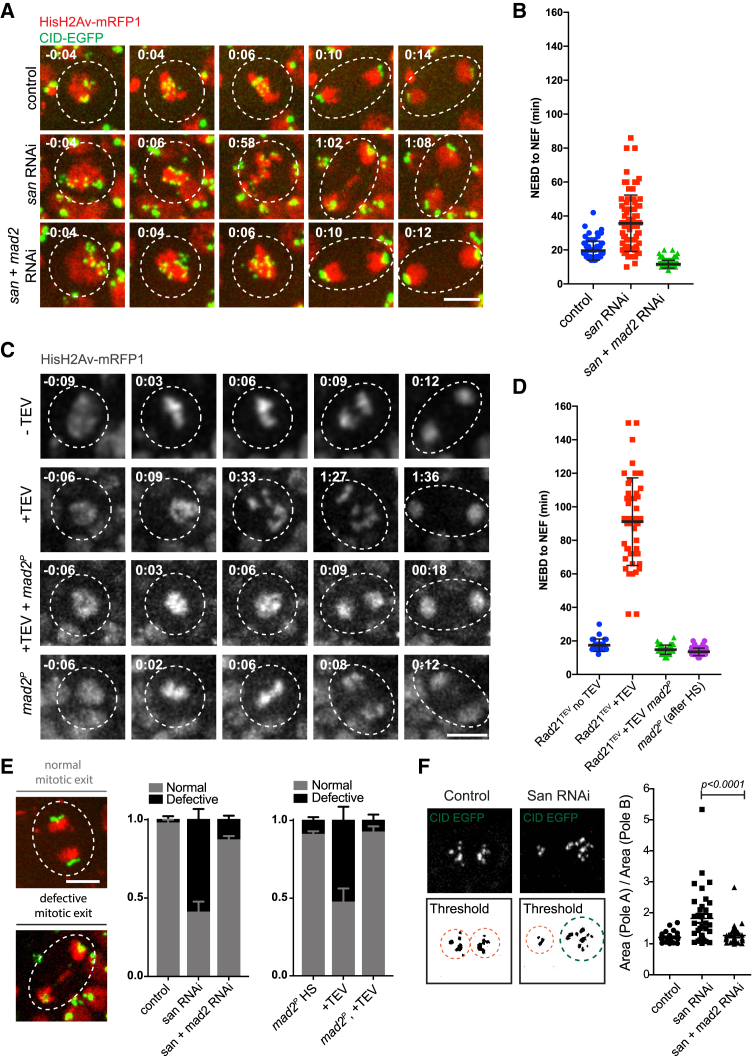
Figure 2.
Inhibition of the SAC in Wing Imaginal Discs Alleviates Mitotic Errors Caused by Premature Loss of Cohesin
(A) Images from movies of the wing disc pouch in the control, san RNAi, and san and mad2 RNAi strains. Strains contained HisH2Av-RFP (red) and Cid-EGFP (green). Times are relative to NEBD. The scale bar represents 5 μm.
(B) Quantification of mitotic duration in control, san RNAi, or san and mad2 RNAi strains. The duration of mitosis was measured from nuclear envelope breakdown (NEBD) to nuclear envelope formation (NEF) using H2Av-RFP channel. Images were taken every 2 min. Each dot represents an individual cell and lines represent mean ± SD (n = 71/5 for control, 77/5 for san RNAi, and 124/5 for san+mad2 RNAi; n = number of cells and number of independent discs).
(C) Images from movies of the wing disc from strains surviving solely on TEV-cleavable Rad21 (Rad21TEV) with and without heat-shock-induced TEV protease cleavage, in strains wild-type or homozygous mutant for the mad2 gene. Strains also expressed HisH2Av-RFP (red) for visualization of mitotic duration and phenotype. Times are relative to NEBD. The scale bar represents 5 μm.
(D) Quantification of mitotic duration of the no TEV control upon TEV-protease-mediated cleavage of Rad21TEV, upon TEV-protease-mediated cleavage of Rad21TEV in a mad2 mutant background, and in a mad2 mutant without cohesin cleavage but after heat shock. The duration of mitosis was measured from NEBD to NEF using H2AvD-mRFP1. Images were taken every 2 or 3 min. Each dot represents an individual cell, and lines represent mean ± SD (n = 27/4 for Rad21TEV − TEV [no heat shock (HS)], 46/8 for Rad21TEV + TEV, 46/4 for Rad21TEV+TEV in a mad2P background, and 60/4 for mad2P after HS; n = number of cells and number of independent discs).
(E) Representative images of mitotic cells from san RNAi undergoing mitosis with normal and defective mitotic exit. The scale bar represents 5 μm and applies to all images. Graph represents the quantification of mitotic defects observed in the different experimental conditions as mean ± SEM of errors of individual discs (n ≥ 4 independent discs corresponding to over 50 cells analyzed per experimental condition).
(F) Quantification of centromere (cid-EGFP-labeled) segregation assymetry in the different experimental conditions; graph represents segregation symmetry index per cell calculated as the area of pole A (with higher area) divided by area of pole B (lower area), as illustrated on the left (n ≥ 4 independent discs corresponding to over 50 cells analyzed per experimental condition). Statistical analysis was performed using one-way ANOVA test.