Figure 1.
TUT4 and TUT7 Restrict L1 Retrotransposition
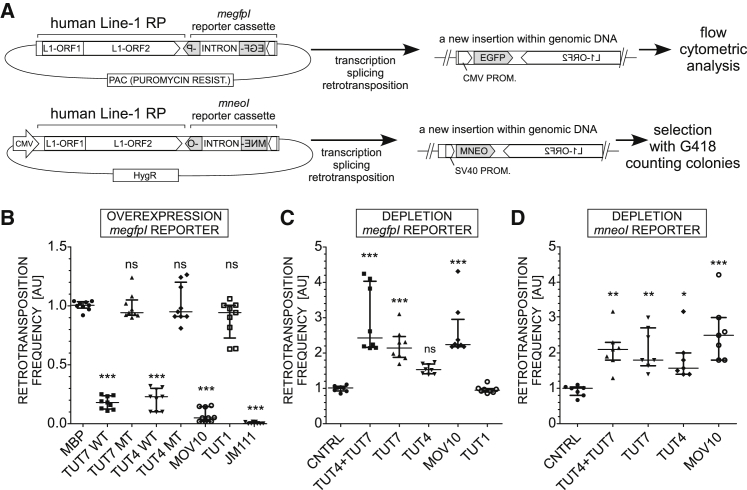
(A) Flowchart of the plasmid-based L1 retrotransposition assays that allow assessment of retrotransposition events by either flow cytometry-based monitoring of cellular EGFP fluorescence (megfpI reporter) or counting drug-resistant colonies (mneoI reporter).
(B) Effects of overexpression of either WT or mutant (MT) TUT4 or TUT7 or WT TUT1, MOV10, or MBP (control) (each point = biological replicate) on L1 retrotransposition in HEK293T cells. Negative control (JM111): a retrotransposition-defective reporter (L1-ORF1pR261A/R262A). The results of independent experiments were normalized relative to the control (MBP). Statistical significance was calculated using one-way ANOVA and Tukey’s multiple comparison test (∗∗∗p < 0.001; ∗∗p < 0.01; and ∗p < 0.05, in comparison to MBP).
(C) Retrotransposition assay in HEK293T cells depleted of TUT7, TUT4 (alone or combined), MOV10, or TUT1 using siRNAs. A control with a non-targeting siRNA was included (CNTRL). Normalization was done relative to CNTRL. Statistical analysis was performed like in (B) (comparison to CNTRL shown). There is no significant difference between CNTRL and TUT1, and a comparison to TUT1 instead of CNTRL gives the same statistical significances.
(D) L1 retrotransposition assays in HeLa-HA cells using mneoI L1 retrotransposition reporter assay. The results were normalized, relative to the control non-targeting siRNA (CNTRL). Statistical analysis is the same as that performed in (B). Shown are comparisons to CNTRL. Normalization was done to CNTRL.
Data in (B)–(D) are represented as medians with individual points and interquartile ranges. See also Figure S1.