Figure 3.
Uridylation of L1 mRNA Abolishes Retrotransposition
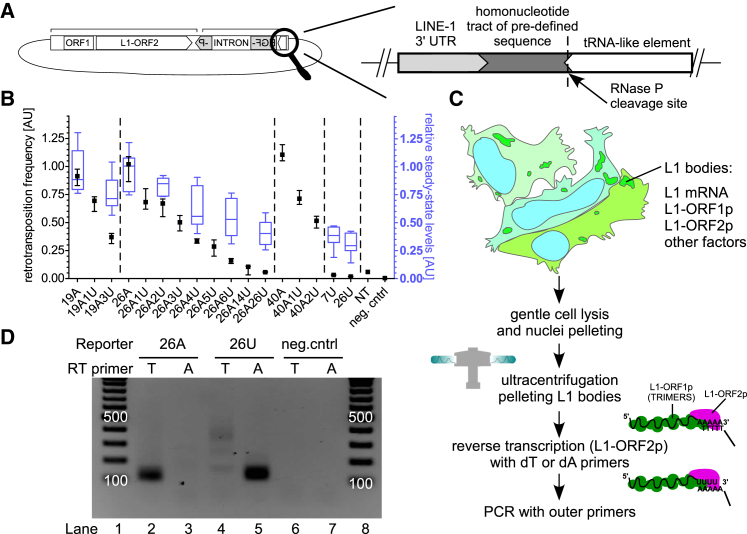
(A) Scheme of the L1 retrotransposition megfpI reporters used in this study. Immediately downstream of a reporter’s 3′ UTR, there is a defined sequence encoding a non-uridylated or differentially uridylated poly(A) (19A, 19A1U, 19A3U, 26A, 26A1U, 26A2U, 26A3U, 26A4U, 26A5U, 26A6U, 26A14U, 26A26U, 40A, 40A1U, 40A2U), 7U, 26U, or the sequence is missing (“no-tail”; NT), all followed by a sequence encoding a tRNA-like element.
(B) Retrotransposition frequency (black) and steady-state reporter mRNA levels (blue) with the reporters described in (A). For retrotransposition assays medians with interquartile ranges are shown (4 to 12 biological replicates). Blue boxes plus whiskers (Tukey’s) represent mRNA abundance (8 biological replicates) for the indicated reporters. Normalizations were done using the 26A reporter. One-way ANOVA and Tukey’s multiple comparison test were used to calculate statistics. All uridylated reporters support significantly (p < 0.001) lower levels of retrotransposition than their non-uridylated counterparts. Steady states: 19A versus 19A3U – ns, 26A versus 26A2U – ns, 26A versus 26A4U/6U/26U, 7U, 26U – p < 0.001.
(C) Scheme of the LEAP procedure with description.
(D) LEAP assays using plasmids carrying LINE-1 reporters ending with a defined sequence (26As or 26Us). The reporter used is indicated at the top, and the RT primer used in each LEAP reaction is indicated below (RT primer). Lanes 1 and 8, a DNA ladder (100 bp to 1,000 bp with 100-bp increments). The 100- and 500-bp bands are labeled. Negative controls (neg.cntrl) without RNPs were also included.
See also Figure S3.