Figure 4.
Differential Effects of TUT4 and TUT7 on L1 mRNA Abundance, Stability, and Translatability
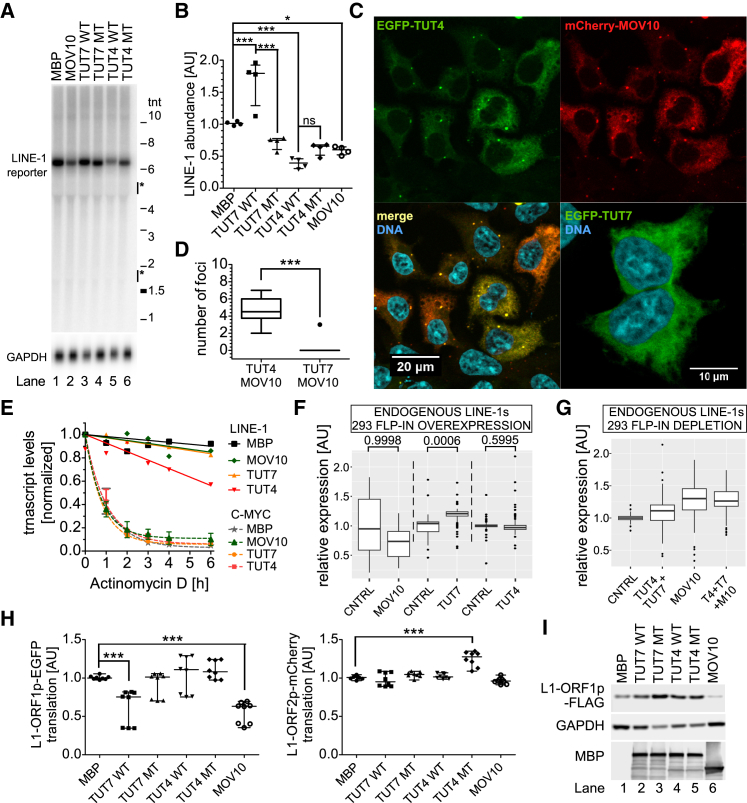
(A) Northern blot of full-length reporter L1 mRNAs, expressed from a plasmid encoding a full-length L1.3 lacking a reporter cassette (JM101/L1.3 no marker) under overexpression of MBP or N’MBP-tagged TUT4, TUT7, and MOV10 as indicated. GAPDH served as a loading control. Marks on the right indicate positions of the RNA reference ladder (in thousands of nucleotides) and the position of 28S and 18S rRNAs is indicated.
(B) Quantification of four northern blots like in (A) (four biological replicates, three independent experiments) normalized relative to the GAPDH signals and the MBP sample. One-way ANOVA and Tukey’s multiple comparison test were used to calculate statistical significance (∗p < 0.05; ∗∗∗p < 0.001).
(C) Confocal microscopy pictures (maximal projections in z) depicting HEK293 cells transfected with plasmids encoding EGFP-TUT4 (top-left panel) and mCherry-MOV10 (top right, merge on bottom-left panel) or EGFP-TUT7 (bottom right) to assess the subcellular localization of proteins. DNA was stained with Hoechst (cyan). Scale bars represent 10 or 20 μm as indicated.
(D) Quantitation of MOV10 containing foci in HEK293 cells that also contain TUT4 or TUT7 (based on co-transfection experiments and confocal microscopy like in C). For each condition (TUT4 vs. TUT7), 30 cells were analyzed. Statistical significance was calculated using a Wilcoxon signed-rank test (p < 0.0001).
(E) Decay of L1 reporter and endogenous MYC mRNAs normalized to GAPDH mRNA. Actinomycin D was added to cell aliquots for 1–6 hr to block transcription, followed by RNA retrieval and estimation of RNA levels by RT-qPCR using multiplexing and Taq-Man probes. Results of three (MYC) or four (L1) independent biological replicates (time-course assays) are shown (mean values).
(F and G) RNA-seq-based estimation of endogenous L1s expressed in HEK293 cells overexpressing TUT4, TUT7 (stable cell lines), or MOV10 (transient transfection) (F) or siRNA-depleted of these proteins (G). Uniquely mapped reads for 76 Homo sapiens-specific L1s (after Repbase) were calculated and normalized to respective controls as indicated. Statistical significances were calculated by DESeq2 for each respective condition pair using summarized counts of each L1 subfamily and are shown above each pair in (F). No significant changes could be observed in (G).
(H) Analytical flow cytometry of cell populations co-transfected with a pJM101/L1.3-O1EGFP-O2mCherry plasmid and plasmids overexpressing the indicated proteins. The pJM101/L1.3-O1EGFP-O2mCherry contains a full-length L1.3 element from which the fluorescent EGFP and mCherry cDNAs were cloned in-frame in the C terminus of L1-ORF1p and L1-ORF2p, respectively. Normalized EGFP and mCherry intensities for data from 8 biological replicates (3 independent experiments) are shown. Statistical significance was calculated like in (B).
(I) Western blot analysis of FLAG-tagged L1-ORF1p, translated from a full-length L1 without a reporter cassette (pZW-L1RP-O1F; Figure S4N). Co-transfected plasmids are indicated at the top of the panel. Membranes were probed with anti-FLAG, anti-GAPDH and anti-MBP antibodies to detect respectively: overexpressed L1-ORF1p-FLAG, GAPDH (loading control), and MBP-tagged proteins. Note that MBP migrates faster than any tagged protein, and it is beyond the blot and thus not detected.
Data in (B), (D), and (H) are medians with individual points and interquartile ranges shown. See also Figure S4 and Tables S3, S4, and S5.