Figure S2.
3′ RACE-Seq of Endogenous and Reporter L1 mRNAs and of Control Cellular mRNAs, Related to Figure 2
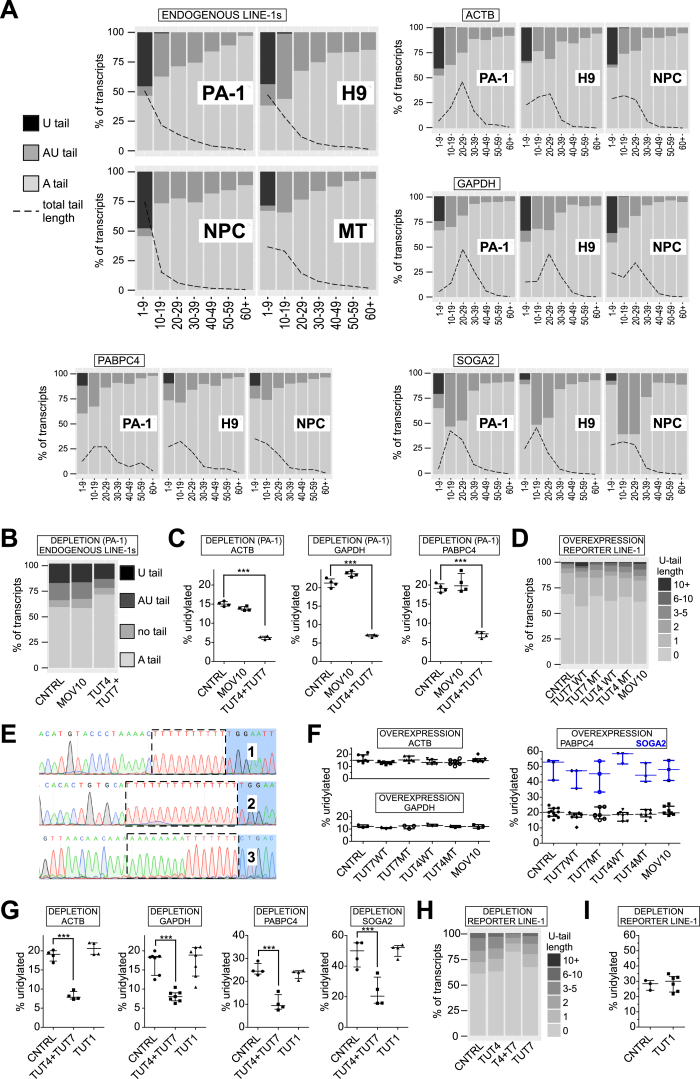
(A) Distribution of U-tails, AU-tails and A-tails in endogenous L1, ACTB, GAPDH, PABPC4 and SOGA2 mRNAs (as indicated) possessing non-templated 3′ end nucleotides in the indicated cells/organs: PA-1 cells, human embryonic stem cells (H9), human neuronal progenitor cells (NPC) and in mouse testes (MT). The fraction of transcript 3′ ends is shown in the y axis with total set to 100%. Tails were binned in 10-nucleotide bins (but 1-9 and 60+) according to their length and are visualized in x axis. A black dashed line overlaid onto the graphs and represents total tail-length distribution, normalized to 100% and shown as % of total transcripts (y axis).
(B) Distribution of 3′ tails in endogenous L1 mRNAs in PA-1 cells transfected with control non-targeting siRNA (CNTRL) or siRNAs against MOV10 or TUT4 and TUT7. The tails were assigned to one of four classes: U-tail (mono- and oligouridylated, but not adenylated); AU-tail (adenylated and mono- and oligouridylated); “no tail” (neither adenylated nor uridylated, mostly truncated within the 3′ UTR); A-tail (oligo- and polyadenylated).
(C) Uridylation of control mRNAs: ACTB, GAPDH and PABPC4 (as indicated) in PA-1 cells transfected with control non-targeting siRNA (CNTRL) or siRNAs against MOV10 or TUT4 and TUT7. Statistical significance was calculated using one-way ANOVA and Tukey’s multiple comparison test significance (∗∗∗p < 0.001). No statistical significance was reported between CNTRL and MOV10 depletion.
(D) Distribution of U-tail lengths in reporter L1 mRNAs in HEK293 cells under overexpression of the indicated proteins. The U-tails were grouped according to the number of uridines. Data were normalized to all mRNAs for a given condition.
(E) Examples of 3′ RACE clones with reporter L1 mRNAs, to show the presence of oligouridylated (1,2) and oligoadenylated and oligouridylated 3′ ends (3). Dashed lined boxes indicate the presence of non-templated nucleotides. Blue background indicates 5′ end of the 3′ adaptor used (different for 1, 2 and 3).
(F) Uridylation of control mRNAs: ACTB, GAPDH, PABPC4 and SOGA2 (as indicated) in HEK293 cells overexpressing MBP (CNTRL), wild-type or mutant TUT4/7 and MOV10. Statistical tests were performed as in panel C. No statistically significant changes were observed.
(G) Uridylation of control mRNAs: ACTB, GAPDH, PABPC4 and SOGA2 (as indicated) in HEK293 cells depleted of TUT4 and TUT7 or TUT1 (as indicated). Statistical significance was calculated as in panel C and is shown where applicable.
(H) Distribution of U-tail lengths on reporter L1 mRNAs in HEK293 cells under depletion of the indicated proteins. The U-tails were grouped according to the number of uridines. Data were normalized to all mRNAs for a given condition.
(I) Uridylation of L1 reporter mRNAs in control HEK293 cells (transfected with non-targeting siRNAs, CNTRL) and in cells depleted of TUT1.
Data on panels C, F, G, and I are represented as medians with individual points and interquartile ranges shown.