Figure 1:
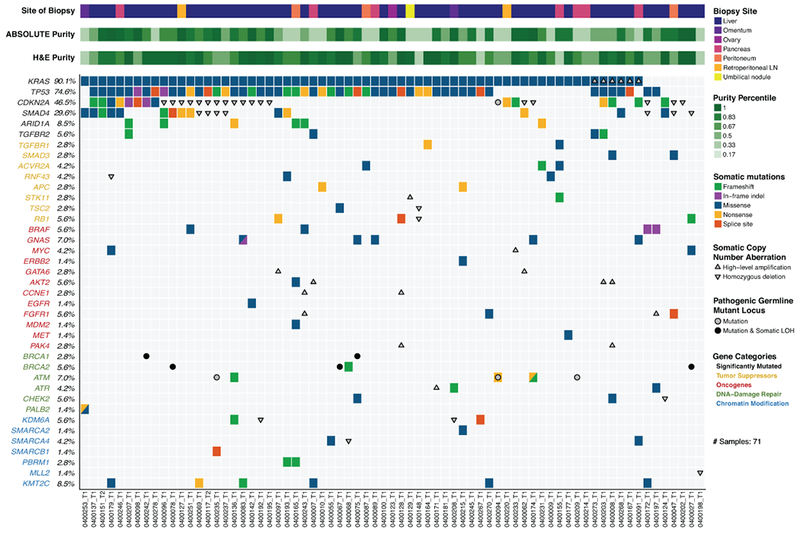
Landscape of genomic alterations identified by whole exome sequencing in biopsies of advanced pancreatic ductal adenocarcinoma (PDAC) patients. Co-mut plot displaying integrated genomic data for 71 samples displayed as columns, including: somatic mutations, high-level amplifications and homozygous deletions, and germline mutations for selected genes. For each sample, the site of biopsy, the ABSOLUTE neoplastic cellularity (purity) from WES data, and the neoplastic cellularity as assessed by histologic evaluation are shown as tracks at the top. Significantly mutated genes with q value ≤ 0.1 that were identified by exome sequencing are listed at the top (black) vertically in order of decreasing significance. Genes from recurrently altered functional classes are also shown, including tumor suppressor genes (yellow), oncogenes (red), DNA damage repair genes (green) and chromatin modification genes (blue). LOH, loss of heterozygosity. LN, lymph node. Indel, insertion/deletion.
