Figure 5:
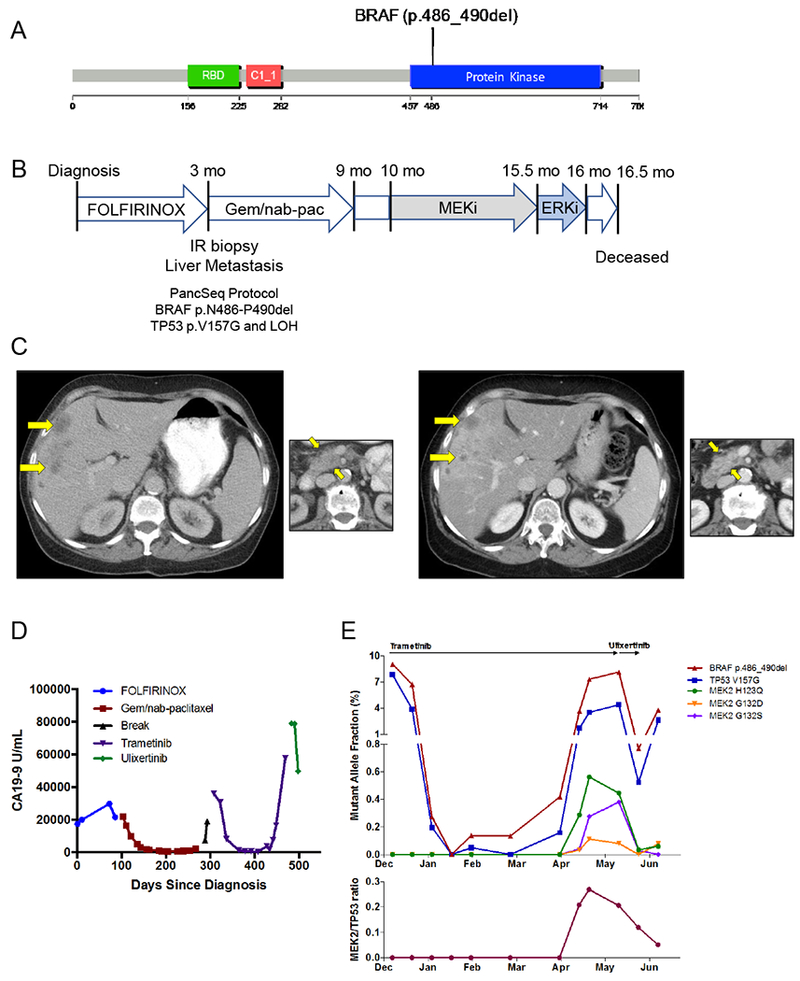
BRAF in-frame deletion confers response to MAPK inhibition. A) An in-frame deletion in BRAF was identified leading to a five amino acid deletion in the kinase domain. B) A 66 year old woman presented with dyspepsia and weight loss and was diagnosed with PDAC and liver metastases and underwent the indicated treatment course. Gem/nab-pac, gemcitabine + nab-paclitaxel. C) CT scans of the liver (large panels) and primary tumor (small panels) at diagnosis (left) and at the time of first restaging scan after 8 weeks of treatment (right) showing partial response to trametinib. D) Serum CA19–9 levels measured throughout the patient’s disease course reflect response and resistance to each therapeutic regimen (color coded by regimen). E) Cell free DNA (cfDNA) measurements for the indicated alleles obtained through droplet digital PCR (ddPCR, BRAF and TP53 alleles) or Guardant360 assay (MEK2 alleles) on plasma collected throughout the patient’s treatment course with trametinib and ulixertinib. Top panel depicts the overall mutant allele fraction of each allele in cfDNA. The bottom panel shows the relative frequency of the MEK2 resistance alleles compared to the overall tumor burden (as measured by the total MEK2/TP53 ratio).
