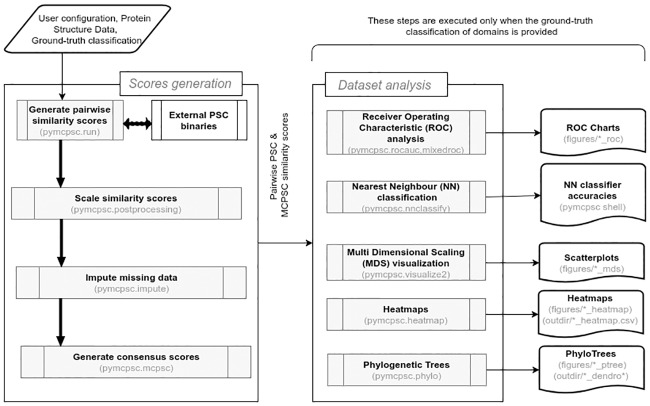
Fig 1. Schematic overview of the architecture of pyMCPSC.
pyMCPSC is organized into several modules, each one implementing a specific functionality. The main entry point of the utility drives the sequence of activities shown. Similarity scores are generated for all protein pairs using the executable binaries of the included PSC methods. Subsequently the scores are scaled, missing data (similarity scores) are imputed and consensus MCPSC scores are calculated for all domain pairs. If the user has supplied ground-truth domain classification information, then comparative analysis results are also generated based on the similarity scores. The modules where the respective functionalities are implemented are specified in parenthesis.