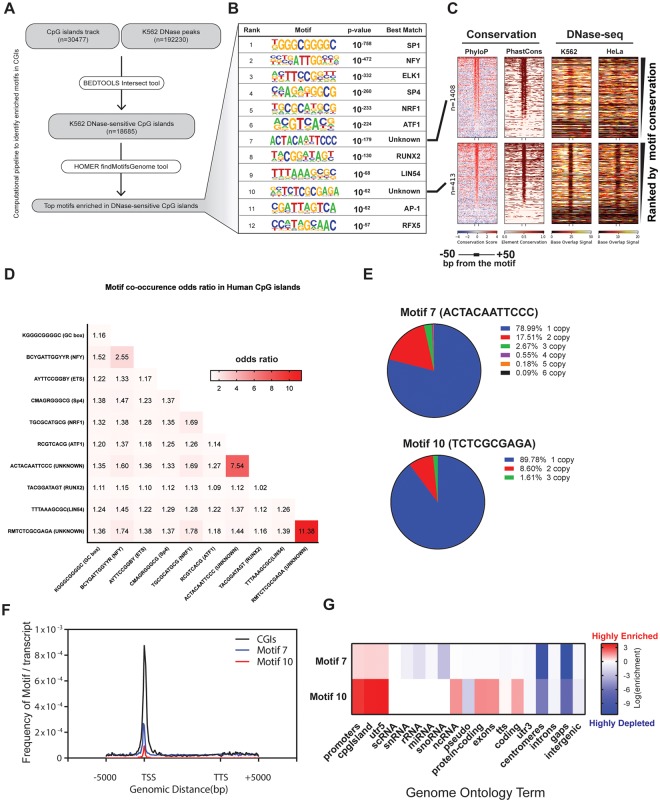
Fig 1. Identification of enriched motifs in human CGIs.
A) The computational pipeline used to identify the most enriched motifs in the DNase-accessible CGIs from K562 cells. Bedtools was used to identify CGIs that overlap with ENCODE derived DNase-sensitive peaks. Homer was then used to identify the most enriched DNA sequence motifs in the DNase-accessible CGIs. B) Known transcription factor binding sites in the enriched motif list. C) Heatmaps showing vertebrate conservation and DNase-seq profiles for motifs #7 and #10 including their flanking 50 bp. PhyloP scores represent conservation of individual base-pairs throughout vertebrate genomes. Positive scores in PhyloP heatmaps, shown by red colors, indicate high sequence conservation whereas negative scores (blue colors) indicate acceleration of base pairs. PhastCons heatmaps show probability scores (ranging from 0 to 1) of conserved DNA elements. DNase-seq heatmaps of K562 and HeLa cells indicate DNase accessibility. Low accessibility, as indicated in dark colors, identifies a central DNase-seq footprint associated with motif #10 in both cell lines. Both motifs occur in DNase-sensitive CGIs of many different cell lines, but only motif #10 was consistently associated with a DNase-seq footprint (Fig A in S1 Fig). D) Motif co-occurrence odds-ratio matrix in DNase-sensitive CGIs. The odds-ratio is the value of observed-to-expected coincidence if motifs were distributed by chance. Higher values indicate a higher likelihood of co-occurrence of indicated motifs in the matrix. E) Pie charts indicate frequency of the motif copy number in human CGIs. F) Metagene profiles for all human CGIs, motifs #7 and #10 were generated by the Homer annotated script, showing that both motifs are associated with TSSs. G) Homer annotated script was used to generate genomic annotation enrichment scores to identify genomic regions where these motifs are enriched. The data indicate that motif #7 and #10 are significantly enriched in promoters and CGIs. The enrichment values were calculated using the cumulative hypergeometric distribution method.