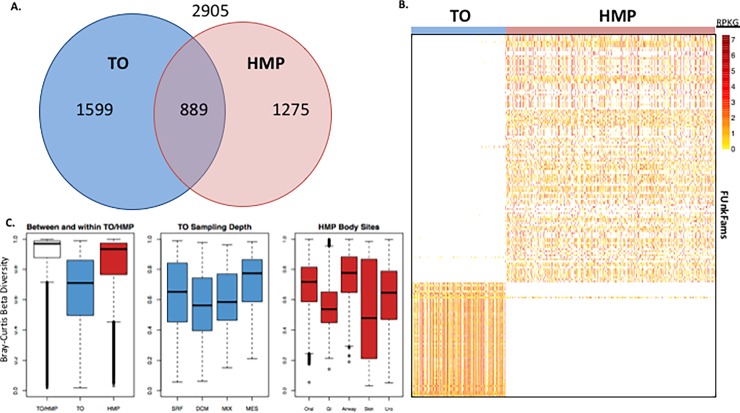
Fig 3. FUnkFams are present in marine and human metagenomes.
(A) Most FUnkFams are detected in either TO or HMP metagenomes (56.6%), but relatively few are present in both environments (13.3%). (B) Heatmap showing the abundance of FUnkFams (rows) in TO (left) or HMP (right) metagenomes after normalizing for gene length, library size, and average genome size (RPKG—reads per kb of gene sequence per genome equivalent [5]). The 180 FUnkFams with at least 50 aligned reads across all samples are displayed (see S4 Fig for the unfiltered heatmap of all FUnkFams). (C) Distributions of Bray-Curtis dissimilarity between pairs of samples from marine environments (TO; blue), between pairs of samples from human microbiomes (HMP; red), and between pairs of samples from different environments (white). Bray-Curtis dissimilarity is a measure of the compositional dissimilarity between two populations, where a value of 1 means the they share no species and 0 means they share all species. Samples are more similar within than between the two environments. SRF, surface water; DCM, deep chlorophyll maximum; MIX, mixed layers; MES, mesopelagic.