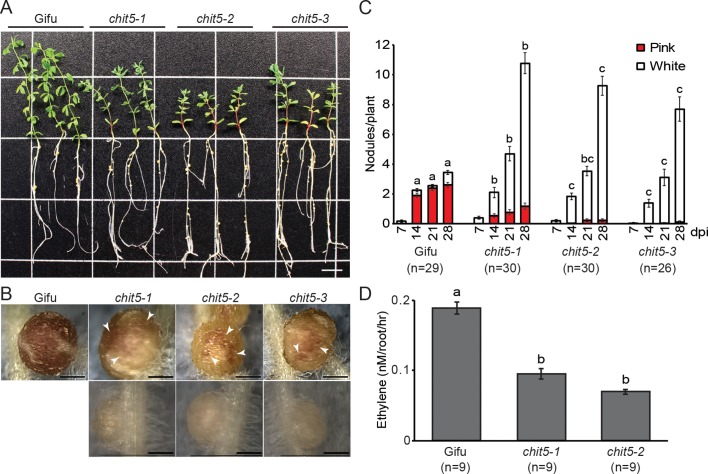
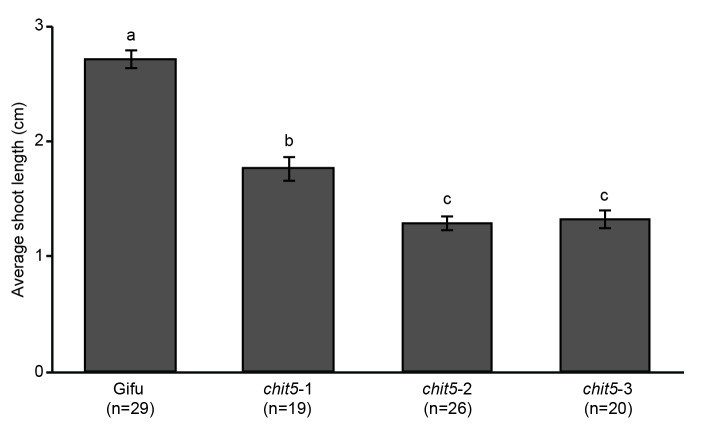
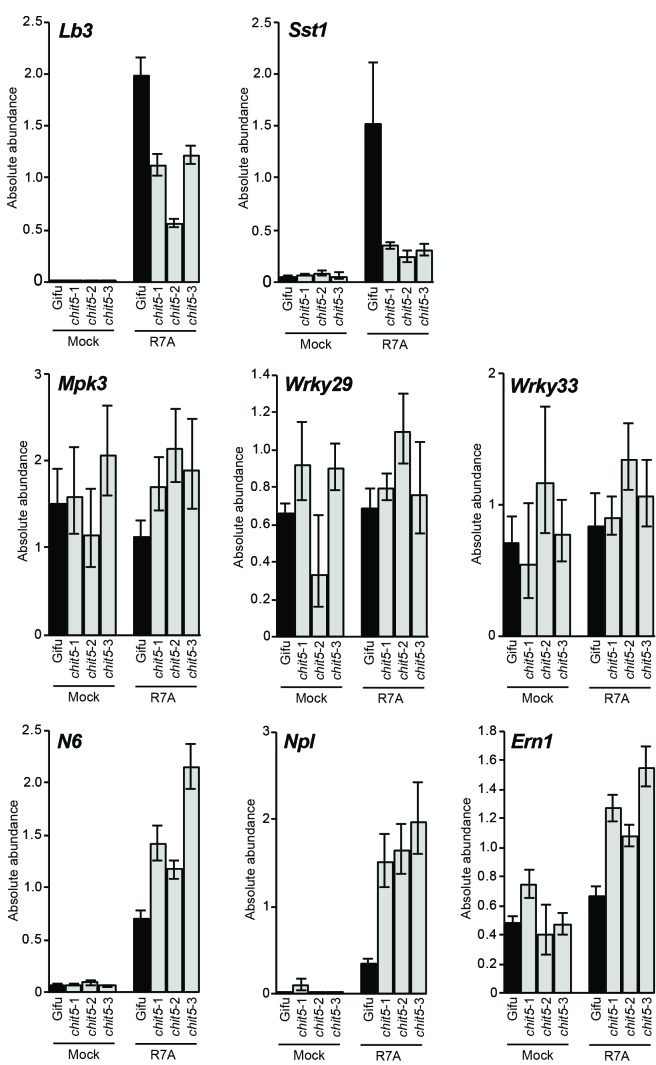
Figure 1. chit5 mutants are defective in nitrogen-fixing symbiosis.
(A) Three representative plants of wild-type Gifu and chit5 mutant alleles 6 weeks post-inoculation with M. loti R7A. Scale bar is 1 cm. (B) Gifu plants form pink nodules whilst chit5 mutants form mainly small white nodules, and occasionally pink-spotted (white arrows) nodules. Scale bars are 0.5 cm. (C) Average number of pink and white nodules formed by M. loti R7A over 4 weeks. (D) Nitrogenase activity measured as the amount of ethylene produced from acetylene in nodules of Gifu and chit5 mutants inoculated with M. loti R7A. (C) and (D) Error bars represent SEM and statistical comparisons of the number of pink nodules formed between genotypes at each time point are shown using ANOVA and Tukey post hoc testing with p values < 0.01 as indicated by different letters.