Abstract
Covalently circularized nanodiscs (cNDs) represent a significant advance in the durability and applicability of nanodisc technology. The new cNDs demonstrate higher size homogeneity and improved stability compared with that of non-circularized forms. Moreover, cNDs can be prepared at various defined sizes up to 80-nm diameter. The large cNDs can house much larger membrane proteins and their complexes than was previously possible with the conventional nanodiscs. In order to experience the full advantages of covalent circularization, high quality circularized scaffold protein and nanodisc samples are needed. Here, we give a concise overview and discuss the technical challenges that needed to be overcome in order to obtain high quality preparations. Furthermore, we review some potential new applications for the cNDs.
Introduction
Phospholipid bilayer nanodiscs offer a detergent-free lipid bilayer model, facilitating the studying of membrane proteins in a physiologically relevant environment[1–3]. A nanodisc is typically composed of two copies of α-helical- amphipathic proteins, termed membrane scaffold proteins (MSPs)[2,3]. The hydrophobic face of MSPs interacts with the lipid acyl chain while the hydrophilic face is located at the outside surface to allow the nanodisc to be soluble in solution. Nanodiscs are widely used for both complexes and single molecule studies of membrane protein structure and function. There are several excellent recent reviews covering nanodisc applications in studying membrane proteins [4•,5•]. While these nanodiscs have been around for over a decade, their utilities for structural and functional studies of membrane proteins have not reached their full potentials. This is due to heterogeneity of size and the number of membrane proteins enclosed, and only small nanodiscs could be constructed with the currently available protein scaffolds[6–8]. To resolve these issues and to expand the stability, applicability and size of nanodiscs, we developed methods to covalently link the N- and C- termini of newly engineered scaffold protein variants based on apolipoprotein A1 (ApoA1) scaffold protein. As a result of the covalent circularization, we produced nanodiscs with a large range of discrete sizes and defined geometric shapes[9••]. Here, we give a concise overview of the technical challenges that needed to be overcome in order to obtain high quality circularized scaffold proteins and nanodiscs. In addition, we discuss the potential new applications that could be offered by these newly engineered nanodiscs.
The benefits of circularization
The linkage of the N- and C-termini of a protein via a peptide bond provides several advantageous properties including improved thermal stability [10–12] and proteolytic resistance [12–16].
Fortunately, both lipid free and lipid-bound ApoA1 have the N- and C- termini in close proximity [17••,18], thus they represent an attractive target for circularization.
Established strategies for circular protein production include the use of various intein-fusion proteins [19,20], which allow circular protein production through expressed-protein ligation or protein trans-splicing. Alternatively, circular proteins can be obtained by using sortase transpeptidases [21], or chemical ligations [22]. More recently, butelase 1 enzyme has been successfully used to circularize peptides and proteins [23].
Sortase A from S. aureus has several advantages that have led to its extensive use for protein site-specific modification and circularization. First, it is easy to get large quantities of the recombinant enzyme. Second, it accepts a broad variety of substrates as long as they contain the LPXTG recognition sequence (X represents any amino acid). Third, circularization using sortase leads to incorporation of only a small five-residue recognition sequence into the circularized product. There are a number of sortase A enzyme variants that can be used for protein circularization. Aside from wild-type sortase A, evolved sortases are available [24,25].
Circularization vs. multimerization of NWs by Sortase A
We have engineered four different variants of circularizable scaffold proteins: NW9, NW11, NW30, and NW50 (NW stands for NanodiscWidth) which assemble ~ 8.5, 11, 15, 50 nm nanodiscs respectively. The scaffold proteins may undergo circularization (intramolecular transpeptidation) or multimerization (intermolecular transpeptidation) followed by circularization[9]. The ratio of circularized product to multimeric product depends both on the size of the protein and on the concentration during the circularization reaction [26].
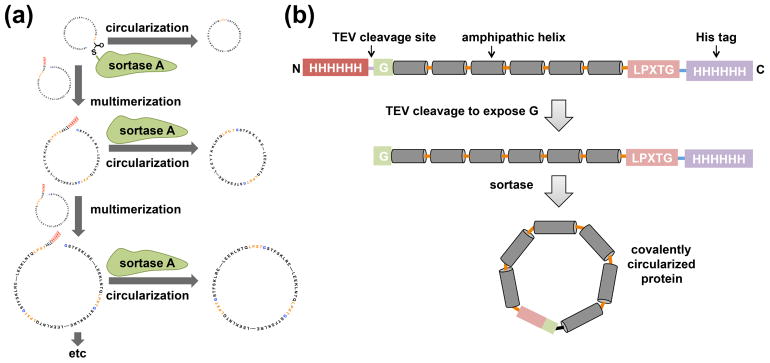
We perform the circularization reaction at lower protein concentration (less than 20 μM) to obtain predominantly monomeric circularized products. Performing the reaction at higher concentration could result in a significant amount of multimerization byproducts (Figure 1a). Interestingly, we found that these multimerization byproducts (circularized) also form nanodiscs that can be separated to some extent by size exclusion chromatography. This observation inspired us to design new DNA constructs for making larger scaffold proteins (i. e NW30 and NW50) [9••].
Figure 1.
Circularization and multimerization of NWs by sortase A. (a) The addition of sortase A to a concentrated scaffold protein solution can lead to multimerization followed by circularization. (b) A general outline of the constructs that are used for making covalently circularized nanodiscs. This figure is adapted from reference 9.
NW constructs expression and purification
All of the NW expression plasmids have been deposited at and are available from the Dana Farber/Harvard Cancer Center (DF/HCC) plasmid depository (http://dnaseq.med.harvard.edu/). The NW constructs contain a tobacco etch virus (TEV) protease-cleavable N-terminal His tag followed by a single glycine, and a C-terminal sortase-cleavable His tag (figure 1b). The presence of these two sites ensures covalent linkage between the N- and C- termini of NW while still preserving the function to form nanodiscs.
The expression yield per liter culture is dependent on the NW construct, media and whether a fermenter or standard shaker was used. In general, the expression of NW9 and NW11 is better than the longer NW variants (NW30 and NW50). We typically obtain around 50–80 mg of purified NW9 and NW11 from 1L LB medium in shaker flasks. On the other hand, we obtain around 7–15 mg for NW30 and only 5 mg for NW50. We observed that a significant amount of the larger NW variants remains in the insoluble fraction during cell lysis. Therefore, we solubilize the insoluble inclusion bodies in guanidine hydrochloride to recover more protein. Table 1 lists some of the issues that can arise during the expression and purification of NW proteins and offers solutions.
Table 1.
Troubleshooting table.
| Problem | Possible reason | Solution |
|---|---|---|
| Expression and purification of NWs | ||
| NW expression yield is low |
|
|
| NW purity is not good after His purification |
|
|
| Removal of the N terminal His tag by TEV is not complete |
|
|
| NW precipitates during TEV cleavage |
|
|
| Circularization of NW | ||
| Circularized reaction is proceeding very slowly or incomplete. |
|
|
| Formation of hydrolysis products |
|
|
NW circularization
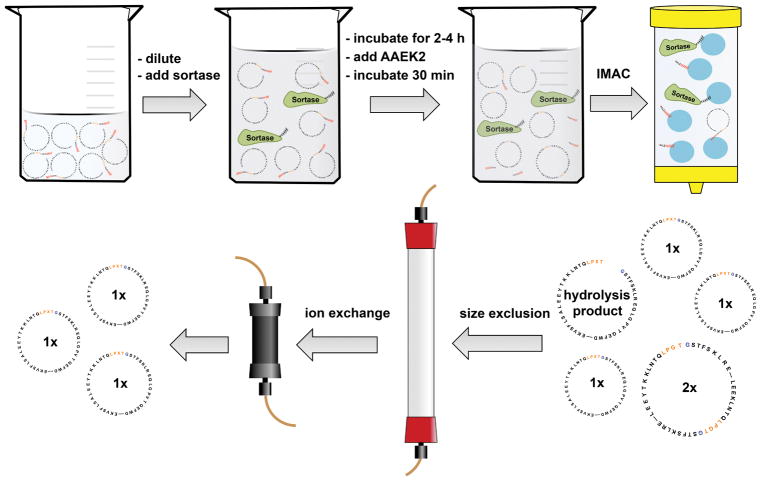
There are three methods that can be used to achieve NW circularization. These methods include circularization over copper chips, over nickel beads, or in solution. We routinely use the in-solution method as we can obtain milligram amounts of circularized proteins. Also, this method is the most cost effective one. We have provided a step-by-step protocol for the production of circularized NW proteins (cNWs) in our earlier paper [9]. The workflow for circularization and the purification is depicted in Figure 2. We dilute the NW to encourage intramolecular circularization and minimize the multimerization reaction. In general, a final NW concentration of 20 uM or less usually resulting in more than 95% circularized monomeric products. We add sortase to a final molar ratio of 2:1, NW: sortase. The reaction is incubated at 37 °C for 2–4 hours. Next, the covalent sortase inhibitor AAEK2 [27] is added, and the solution is incubated for another 30 min at room temperature. Protein that did not undergo circularization is removed by binding to a Ni-NTA column. The cNWs are further purified by size-exclusion chromatography to remove most of the multimerization products, followed by ion exchange chromatography to eliminate the hydrolysis products and the remaining multimerization products.
Figure 2.
Schematic of the in solution method for scaffold protein circularization. After the IMAC step, cNWs are further purified by size-exclusion followed by ion exchange chromatography to remove the multimerization and hydrolysis byproducts.
Circularized nanodisc assembly: General considerations
There are several factors to consider when attempting incorporation of a membrane protein into cNDs. First, the lipid to NW ratio is very important and needs to be empirically determined in each membrane protein case in order to obtain a high yield of incorporation and a homogenous nanodisc preparation. This can be done by screening for nanodisc homogeneity and yield by size exclusion chromatography. Depending on the required surface area of a membrane protein that needs to be inserted, we subtract the corresponding number of lipid molecules from the initial lipid to NW ratio. We estimate that a single transmembrane (TM) helix usually takes up around 70–80 Å2. Thus, around 560 Å2 would be required for a 7 TM-helical protein like a GPCR this would require around.
Using a suboptimal lipid amount will result in loose and unstable nanodiscs as the lateral pressure imparted by the belt protein will be abolished or significantly reduced. In this case, the lipid will be less constrained and the movement of the embedded membrane protein will not be reduced. On the other hand, using excess lipids will not be as damaging since the covalent circularization of the belt protein will not allow the nanodisc to exceed the belt’s maximum diameter. In this case, the lateral pressure imparted by the belt protein will be fully utilized. One potential issue that could arise when using excess lipids is the formation of lipid aggregates and liposomes. These aggregates/liposomes are easy to separate from the smaller nanodiscs (9,11,15 nm nanodiscs), however it might become challenging to separate them from the larger ones (i.e 50 nm nanodiscs).
It is important to identify the ideal lipid composition that supports function/stability for a particular membrane protein. Often we find that choosing the ideal lipids leads to better incorporation yield.
The smallest nanodiscs (8.5, 11 nm) are the most stable and thus are well suited for NMR structural and dynamic studies. The larger size nanodiscs still provide long-term stability of the incorporated membrane protein and allow for high resolution structural studies by cryo-EM.
Potential new opportunities offered by covalently circularized nanodiscs
1- Better Vaccine
Conventional nanodiscs have shown promise in vaccine development. Several antigens including the viral protein gp160 from HIV [30] and hemagglutinin (HA) from influenza [31] have been successfully incorporated into nanodiscs. HA in nanodisc (HA-ND) vaccination induced efficient immune response and protective effect in a mouse model. Sera from HA-ND-immunized mice had significantly higher anti-HA IgG levels than HA-only immunized mice. Moreover, HA-ND induced anti-HA antibody response that was more broadly neutralizing than that induced by HA alone. Recently, ApoA1-mimetic peptide based nanodiscs have been used to develop a new vaccine system suited for personalized cancer immunotherapy [32••].
The improved stability of cND relative to conventional nanodiscs not only could have impact on the shelf life of the vaccine preparations but also could improve the antigen lifetime in vivo, which might be essential in some cases. Furthermore, cNDs could provide improved homogeneity and better control of the number of antigens incorporated into each nanodisc. Finally, the larger size cNDs could be very useful to incorporate several copies of antigens/adjuvents in the same nanodisc.
2- Structural and functional studies of large membrane protein complexes in bilayer
Large cNDs provide tools to incorporate much larger membrane proteins or their intramembrane and extramembrane complexes than previous nanodisc systems have allowed. For example, these large nanodiscs could be used to incorporate and study the various pore architectures and the membrane insertion mechanisms of pore forming toxins and proteins (PFPs). The structural studies of these PFPs could provide insights to enable the development of novel therapeutic strategies that would prevent the development of bacterial resistance. Also, large cNDs can be used to study very large complexes such as mammalian respiratory complex I which contains 45 subunits and represents one of the largest membrane-bound enzymes in the cell [33].
3- Virus entry
One of the most exciting new applications of large cND is providing enough surface area to act as a surrogate membrane for the study of the early steps in virus infection. For example, we have used 50-nm cNDs to study the question of how simple non-enveloped viruses like poliovirus (~ 30 nm) transfer their genomes across membranes to initiate infection. EM images show the poliovirus bound to 50-nm cNDs, which are decorated with the CD155 receptors. Excitingly, the EM images reveal the formation of a putative pore in the cNDs and show individual viruses ejecting RNA across membranes [9].
Currently, we are working on extending the nanodisc sizes beyond 80 nm for two reasons; first, we would like to study the viral entry of larger non-enveloped viruses. Second, we believe these very large nanodiscs (> 80 nm) will be useful in studying enveloped virus binding and membrane fusion.
Conclusions
Covalently circularized nanodiscs represent a significant advance in the durability of nanodisc technology. We discussed the technical challenges that need to be overcome to obtain high quality cND preparations and offered solutions. Furthermore, we outlined some potential new applications that could be offered by these newly engineered nanodiscs.
Highlights.
cNDs represent a significant advance in the applicability and durability of nanodisc technology.
We highlighted the technical challenges that needed to be overcome to obtain high quality cNDs.
Large cNDs can be used to study the early steps in virus infection.
Acknowledgments
Funding
This work was supported by NIH grant F32GM113406 to M.L.N and grants: GM047467 and AI037581 to G. W.
Footnotes
Conflict of Interest.
The authors have founded NOW Scientific Inc to allow purchase of assembled circularized MSPs. Expression plasmid are available for a nominal fee, and detailed protocols have been published.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• of outstanding interest
- 1.Bayburt TH, Carlson JW, Sligar SG. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J Struct Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 2.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters. 2002;2:853–856. [Google Scholar]
- 3.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •4.Denisov IG, Sligar SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. This comperhensive review article summarizes the applications of nanodisc technology. This article cites over 500 publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •5.Rouck JE, Krapf JE, Roy J, Huff HC, Das A. Recent advances in nanodisc technology for membrane protein studies (2012–2017) FEBS Lett. 2017;591:2057–2088. doi: 10.1002/1873-3468.12706. This review article highlights techniques that have been recently combined with nanodisc technology to study membrane protein from 2012–2017. The authors evaluate the advantages and shortcomings of using nanodiscs in the study of membrane proteins. This article cites over 200 publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raschle T, Hiller S, Yu TY, Rice AJ, Walz T, Wagner G. Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grinkova YV, Denisov IG, Sligar SG. Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel. 2010;23:843–848. doi: 10.1093/protein/gzq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods. 2017;14:49–52. doi: 10.1038/nmeth.4079. The authors engineered covalently circularized nanodiscs with high homogeneity in size and with improved stability compared with that of non-circularized ones. •• They used 50-nm cND as a model membrane for studying the poliovirus interactions with a membrane. EM images show the formation of a putative pore in the cND. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens CA, Semrau J, Chiriac D, Litschko M, Campbell RL, Langelaan DN, Smith SP, Davies PL, Allingham JS. Peptide backbone circularization enhances antifreeze protein thermostability. Protein Sci. 2017;26:1932–1941. doi: 10.1002/pro.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trabi M, Craik DJ. Circular proteins--no end in sight. Trends Biochem Sci. 2002;27:132–138. doi: 10.1016/s0968-0004(02)02057-1. [DOI] [PubMed] [Google Scholar]
- 12.Iwai H, Pluckthun A. Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999;459:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 13.Qi X, Xiong S. Intein-mediated backbone cyclization of VP1 protein enhanced protection of CVB3-induced viral myocarditis. Sci Rep. 2017;7:41485. doi: 10.1038/srep41485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popp MW, Ploegh HL. Making and breaking peptide bonds: protein engineering using sortase. Angew Chem Int Ed Engl. 2011;50:5024–5032. doi: 10.1002/anie.201008267. [DOI] [PubMed] [Google Scholar]
- 15.Borra R, Camarero JA. Recombinant expression of backbone-cyclized polypeptides. Biopolymers. 2013;100:502–509. doi: 10.1002/bip.22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL. Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci U S A. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••17.Bibow S, Polyhach Y, Eichmann C, Chi CN, Kowal J, Albiez S, McLeod RA, Stahlberg H, Jeschke G, Guntert P, et al. Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat Struct Mol Biol. 2017;24:187–193. doi: 10.1038/nsmb.3345. The authors solved the structure of a lipid bound MSP by usign a combination of NMR, EPR and TEM data. The MSPs arrange as double belt that surrounds a lipid bilayer patch in an antiparallel fashion. The structure give important insights into the dimerization and lipid interactions of MSP. [DOI] [PubMed] [Google Scholar]
- 18.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc Natl Acad Sci U S A. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Elleuche S, Poggeler S. Inteins, valuable genetic elements in molecular biology and biotechnology. Appl Microbiol Biotechnol. 2010;87:479–489. doi: 10.1007/s00253-010-2628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans TC, Jr, Benner J, Xu MQ. The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. J Biol Chem. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 21.Antos JM, Popp MW, Ernst R, Chew GL, Spooner E, Ploegh HL. A straight path to circular proteins. J Biol Chem. 2009;284:16028–16036. doi: 10.1074/jbc.M901752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowper B, Craik DJ, Macmillan D. Making ends meet: chemically mediated circularization of recombinant proteins. Chembiochem. 2013;14:809–812. doi: 10.1002/cbic.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol. 2014;10:732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 24.Dorr BM, Ham HO, An C, Chaikof EL, Liu DR. Reprogramming the specificity of sortase enzymes. Proc Natl Acad Sci U S A. 2014;111:13343–13348. doi: 10.1073/pnas.1411179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc Natl Acad Sci U S A. 2011;108:11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z, Guo X, Guo Z. Sortase A-catalyzed peptide cyclization for the synthesis of macrocyclic peptides and glycopeptides. Chem Commun (Camb) 2011;47:9218–9220. doi: 10.1039/c1cc13322e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maresso AW, Wu R, Kern JW, Zhang R, Janik D, Missiakas DM, Duban ME, Joachimiak A, Schneewind O. Activation of inhibitors by sortase triggers irreversible modification of the active site. J Biol Chem. 2007;282:23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 29.Lucast LJ, Batey RT, Doudna JA. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques. 2001;30:544–546. 548, 550. doi: 10.2144/01303st06. passim. [DOI] [PubMed] [Google Scholar]
- 30.Nakatani-Webster E, Hu SL, Atkins WM, Catalano CE. Assembly and characterization of gp160-nanodiscs: A new platform for biochemical characterization of HIV envelope spikes. J Virol Methods. 2015;226:15–24. doi: 10.1016/j.jviromet.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya P, Grimme S, Ganesh B, Gopisetty A, Sheng JR, Martinez O, Jayarama S, Artinger M, Meriggioli M, Prabhakar BS. Nanodisc-incorporated hemagglutinin provides protective immunity against influenza virus infection. J Virol. 2010;84:361–371. doi: 10.1128/JVI.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••32.Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater. 2017;16:489–496. doi: 10.1038/nmat4822. The authors designed and used ApoA1-mimetic peptide based nanodiscs to develop a new vaccine system for personalized cancer immunotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]