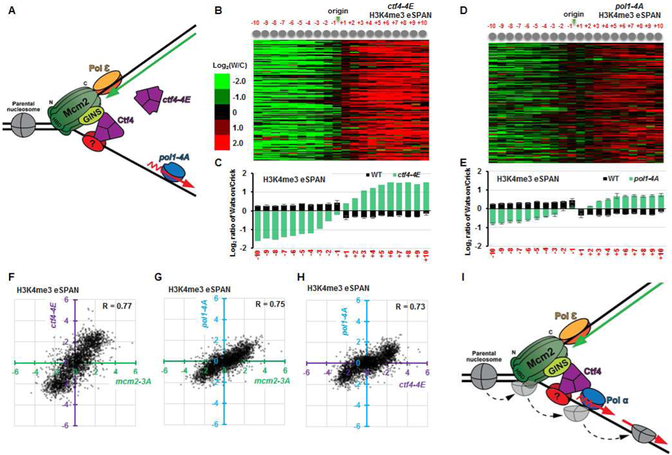
Figure 6. Disruption of the CMG-Ctf4-Pol1 interaction impairs the transfer of parental histone (H3-H4)2 to lagging strands.
(A) Mutation of the Ctf4-interacting-peptide of Pol1 (pol1-4A) or the Ctf4 mutant defective in binding to both CMG and Pol1 (ctf4-4E) uncouples the CMG helicase from the Polα primase at lagging strand. (B-E) Analysis of the average bias pattern of H3K4me3 eSPAN peaks in ctf4-4E (B-C) or pol1-4A (D-E) mutant cells. The leading strand bias ratio of H3K4me3 eSPAN in pol1-4A cells is smaller than that of mcm2-3A, ctf4Δ, or ctf4-4E cells. (F-H) The dot scatter plots show the H3K4me3 bias pattern in ctf4-4E vs mcm2-3A (F), pol1-4A vs mcm2-3A (G), andpol1-4A vs ctf4-4E (H). Each dot represents bias ratio of H3K4me3 eSPAN peaks at a single nucleosome among 134 early DNA replication origins (from −10 to +10, n=2680). (I) Model depicting how the MCM helicase and Pol1, connected by Ctf4, regulate the transfer of parental (H3-H4)2 to the lagging strand. See also Figures S5 and S6.