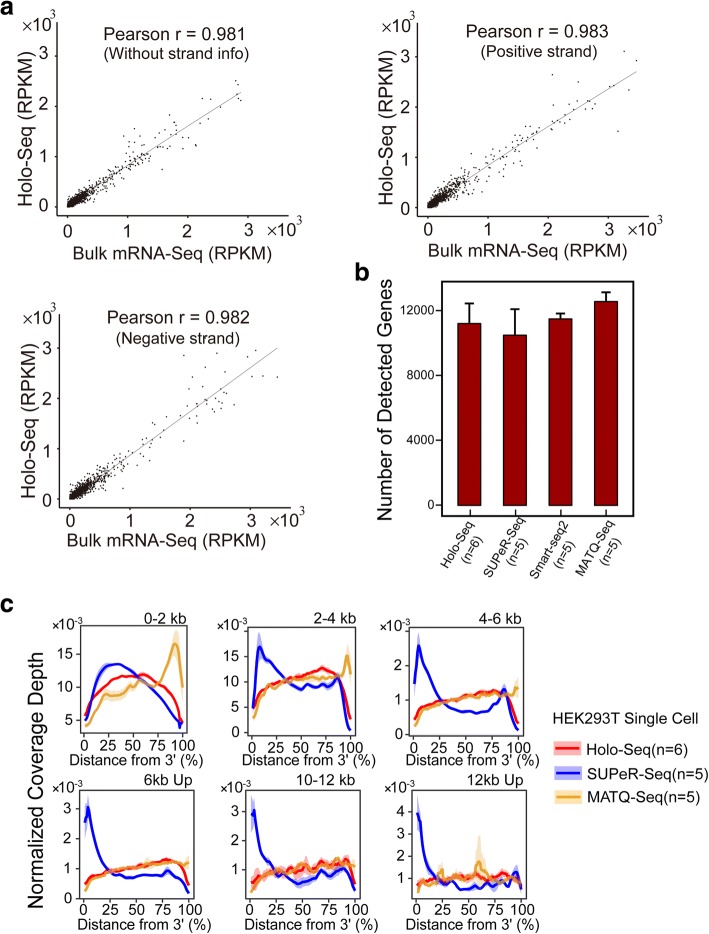
Fig. 2.
Holo-Seq accurately profiles total RNAs with a complete strand of origin information from single cells. a RPKM scatterplots of expressed genes between the combined dataset (total RNA with a complete strand of origin information from 10 mESCs single cells) and a directional bulk mRNA-Seq. b Comparison of the detected gene number in HEK293T single cells at the maximum exome-mapped depth of MATQ-Seq (UMI labeled reads) and 1.2M unique exome-mapped depth of Holo-Seq, SUPeR-Seq, and Smart-Seq2. c Read coverage across transcripts of different lengths of three methods in HEK293T single cells. The read coverage over the transcripts is displayed along with the percentage of the distance from their 3′ end. Shaded regions indicate the standard deviation