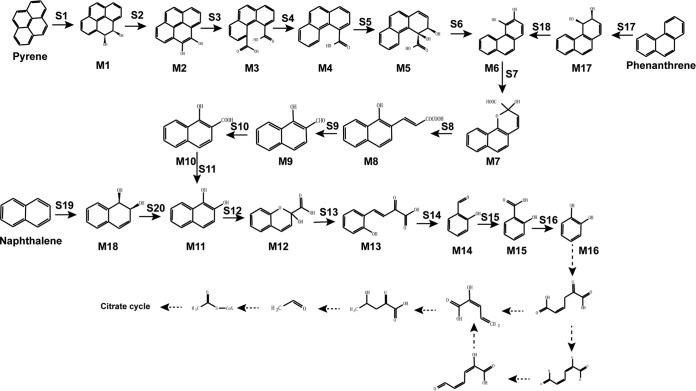
FIG 6.
Complete PAH degradation pathway in Cycloclasticus sp. P1 based on genomic, transcriptomic, and other experimental analyses. Chemical designations: M1, pyrene-cis-4,5-dihydrodiol; M2, 4,5-dihydroxypyrene; M3, phenanthrene-4,5-dicarboxylic acid; M4, phenanthrene-4-carboxylate; M5, cis-3,4-dihydroxyphenanthrene-4-carboxylate; M6, 3,4-dihydroxyphenanthrene; M7, 2-hydroxy-2H-benzo[h]chromene-2-carboxylic acid; M8, trans-4-(1′-hydroxynapth-2-yl)-2-oxobut-3-enoic acid; M9, 1-hydroxy-2-naphthaldehyde; M10, 1-hydroxy-2-naphthoic acid; M11, 1,2-dihydroxynaphthalene; M12, 2-hydroxy-2H-chromene-2-carboxylic acid; M13, trans-o-hydroxybenzylidenepyruvic acid; M14, salicylaldehyde; M15, salicylic acid; M16, catechol; M17, cis-3,4-phenanthrenedihydrodiol; M18, (1R,2S)cis-l,2-naphthalenedihydrodiol. Enzyme designations: S1, pyrene dioxygenase (RHD-1); S2 and S6, dihydrodiol dehydrogenase (Q91_0872); S3, S4, S7, and S12, cleavage dioxygenase (Q91_2224); S5, dioxygenase (RHD-4); S8 and S13, isomerase (Q91_2218); S9 and S14, hydratase-aldolase (Q91_0437); S10, aldehyde dehydrogenase (Q91_0507); S11, 1-hydroxy-2-naphthoate hydroxylase (Q91_0344); S15, salicylaldehyde dehydrogenase (Q91_0488); S16, salicylate 1-hydroxylase (Q91_1959); S17, phenanthrene dioxygenase (RHD-2); S18, dihydrodiol dehydrogenase (Q91_2235); S19, naphthalene 1,2-dioxygenase (RHD-3); S20, NAD+-dependent cis-1,2-naphthalenedihydrodiol dehydrogenase (Q91_2228). Known metabolic pathways are represented by solid arrows. Predicted metabolic pathways are represented by dotted arrows.