Figure 1. Illustration of the mutations introduced into the putative 5’ RmRE along with the MMTV transcripts expressed using specific splice sites.
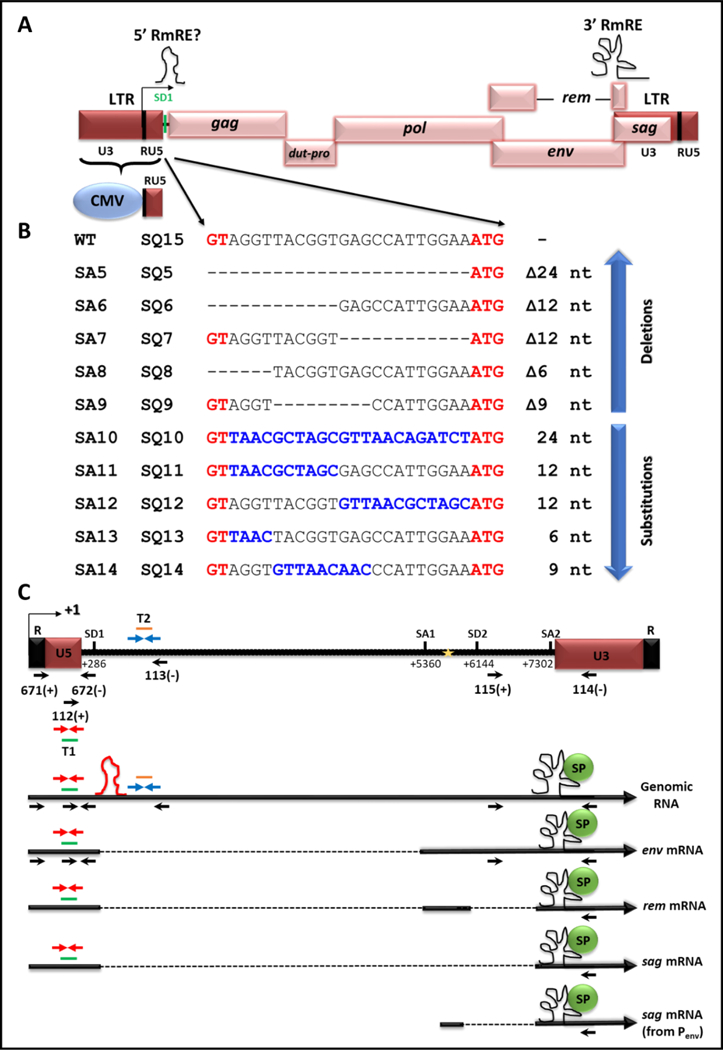
(A) Schematic representation of the MMTV genome with and without the hybrid (CMV/R/U5) LTR. Locations of the 3’ RmRE as well as that of the putative 5’ RmRE are demarcated on the genome with schematics of higher order structures. (B) Depiction of mutations introduced between SD1 and gag start codon (ATG) and cloned into the full length wild type (WT) clone, HYB-MTV (SA series of clones), or cloned into MMTV with the hybrid hCMV/R/U5 5’ LTR (SQ series of clones). The sequences in black represent the WT sequences, whereas the sequences in red represent the SD1 (GT) and gag initiation codon (ATG). Dashes represent the deleted sequences, whereas sequences in blue denote substituted sequences. (C) Illustration of MMTV gRNA followed by the major transcripts produced from the use of the indicated splice sites. The mapped primers show the location of common primers used in this study. For the sake of clarity, the words OTR and OFM have been omitted from the primer names, showing only primer numbers (see Table 1 for primer sequences). SD1, major splice donor; SD2, splice donor in the env gene. Gold star; the internal promoter in the env gene. The red stem loop depicts the location of the putative 5’ cis-acting element. The black structural element depicts the 3’ Rem responsive element (RmRE). T1, Taqman Assay 1 used to quantitate all MMTV mRNAs; T2, Taqman Assay 2 used to quantitate the full-length gRNA.
