Figure 2: A region between SD1 and the gag ATG is important for MMTV mRNA expression.
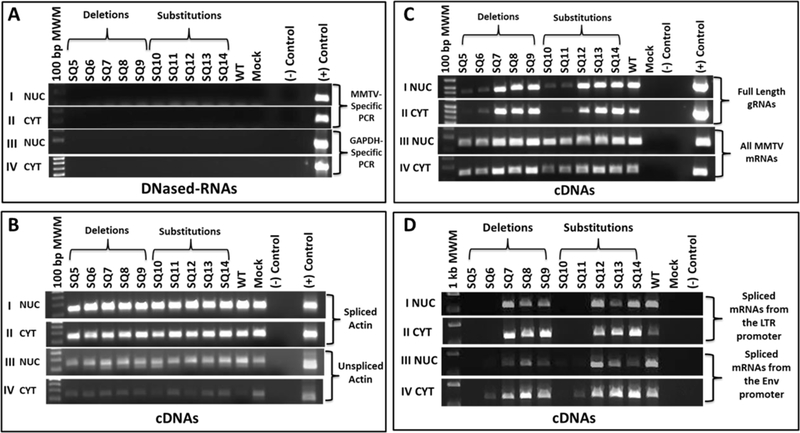
(A) DNase-treatment of nuclear (NUC) and cytoplasmic (CYT) RNAs were tested by PCR. Panels I & II: MMTV-specific primers (OTR671/OTR672) for plasmid DNA contamination; Panels III & IV: Cellular GAPDH primers for genomic DNA contamination. (B) RT-PCR amplification of spliced and unspliced ß-actin mRNA as a subcellular-fractionation control. Panels I & II: Spliced mRNA amplification of ß-actin in NUC and CYT fractions, respectively. Panels III & IV: Unspliced mRNA amplification of ß-actin in NuC and CYT cDNAs. (C) RT-PCR analysis of: Panels I & II, full-length (OFM112/OFM113) and Panels III & IV, all MMTV mRNAs (OTR671/OTR672) in NUC and CYT cDNA fractions. (D) RT-PCR analysis of other MMTV mRNAs using: Panels I & II, OFM112/OFM114 primers that detect spliced mRNAs from the LTR promoter. Expected sizes: env 2485 bp/rem 1324 bp/sag 542 bp. The band corresponding to sag mRNA is shown since the other bands could not be detected consistently; Panels III & IV: OFM115/OFM114 primers that detect spliced mRNAs from the env promoter. Expected sizes: env 1728 bp/rem and sag 567 bp. Once again, only the 567 bp band could be detected under the PCR conditions used.
