Figure 5: RNA stability analysis of 5’ UTR mutants.
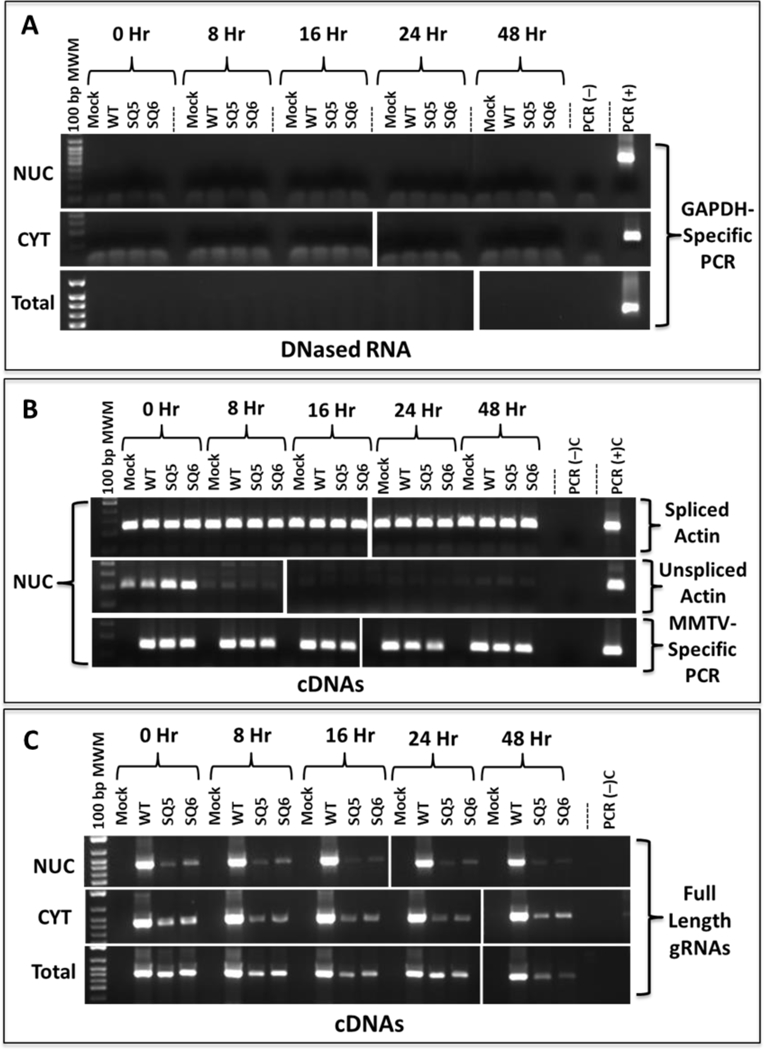
HEK293T cells transfected with mutant and wild type (WT) MMTV constructs and treated with Actinomycin D for various time points. Nuclear (NUC), cytoplasmic (CYT), and total extracted RNAs were DNase-treated and analyzed by RT-PCR for: (A) Lack of DNA contamination in Panels I, NUC, II, CYT, and III, Total RNA samples using GAPDH-specific primers. (B) Panel I, spliced B-actin (247 bp), panel II, unspliced ß-actin (203 bp), as well as panel III, all MMTV mRNAs (142 bp) in the NUC fractions using primers OTR671 and OTR672. (C) MMTV full-length genomic RNAs (723 bp) in panel I, NUC, panel II, CYT, and panel III, total RNA using primer pair OFM112/OFM113. Mock, HEK293T cells transfected with pcDNA3 plasmid alone.
