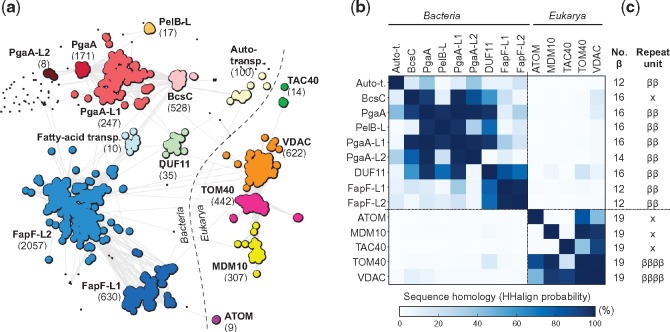
Fig. 2.
—Classification and HMM-comparison of bacterial and mitochondrial OMBBs (MOMBBs). (a) Cluster map of a total of 5277 sequences collected for each MOMBB family and bacterial FapF-like and PgaA-like OMBBs. Clustering was performed with CLANS in 2D until equilibrium at a BLASTp P-value of 1.0. Connections represent similarities up to a P-value of 10−3 (darker means more similar). Black points represent sequences that do not connect to any cluster at P-values <10−4. The number of sequences in each cluster is shown within brackets. Clusters composed solely by hypothetical and nonannotated sequences but with significant homology to a known protein family, as detected with HHPred and PSI-BLAST searches, are referred by the name of the homologous family followed by Lx, where x represents the number of the cluster. The taxonomic distribution of the collected sequences is illustrated in supplementary figures 1 and 2, Supplementary Material online. A total of five eukaryotic and eight bacterial clusters were obtained. VDAC, TOM40, and MDM10 form a highly connected supercluster, which connects only marginally with bacterial OMBBs. TAC40 connects to VDAC, but ATOM does not connect to any cluster at a P-value <10−3. (b) Sequence homology matrix of OMBB clusters as measured by the hhalign probability of the alignment of their HMM-profiles. Those corresponding to the “Fatty-acid transporters” cluster were not included due to the high level of fragmentation of the sequences composing it. Bacterial and eukaryotic OMBBs define two different regions and all MOMBBs find only marginal matches to bacterial OMBBs, especially BcsC and PgaA, suggesting that all MOMBBs are monophyletic and share only local sequence similarity to OMBBs. (c) Strand composition predicted with Quick2D and repeat units identified with HHrepID for the HMM-profile consensus sequence. All MOMBBs are predicted to have a 19-stranded topology; additionally, VDAC and TOM40 show a repetitive sequence. No bacterial OMBB shows the same topology and repetition pattern. ββ: ββ-hairpin; ββββ: double ββ-hairpin; x: none or not clear.