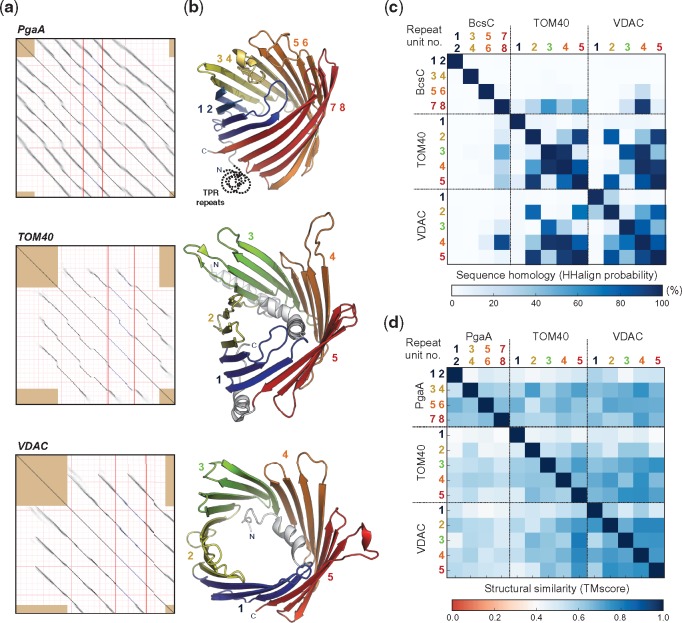
Fig. 3.
—The repetitive nature of VDAC, TOM40, and BcsC/PgaA OMBBs. (a) Self-comparison dot plot of the PgaA, TOM40, and VDAC HMM-consensus sequence generated by HHrepID. The presence of diagonal lines indicates a repetitive sequence. Repeat families were identified at a P-value threshold of 10−1. For VDAC and TOM40, the full consensus sequence included the N-terminal helix, colored grey in (b). Eight sequence repeats were identified in PgaA, whereas only five were found in VDAC and TOM40. (b) Three-dimensional mapping of the identified repeats on their reference three-dimensional structures (PgaA: 4y25_A; TOM40: 5o8o_A; VDAC: 4c69_X). The sequence repeats in PgaA correspond to single ββ-hairpins, while those of VDAC and TOM40 correspond to two ββ-hairpins. (c) Sequence homology matrix, measured as the hhalign probability of the alignment of the HMM-profiles built for VDAC, TOM40, and BcsC (as mapped over PgaA) double ββ-hairpins. The repeats in VDAC and TOM40 find significant matches only with the last C-terminal double ββ-hairpin of BcsC, with the best match found between this and the fourth repeat of VDAC. (d) Structural similarity matrix, measured as the TMscore from structural alignments with TMalign, of VDAC, TOM40, and PgaA double ββ-hairpins. A TMscore below 0.3 indicates random structural similarity while values above 0.5 suggests that both structures assume the same fold. A TMscore of 1.0 denotes a perfect match between the two structures. The predominantly blue matrix suggests that, despite their low sequence similarity, all double ββ-hairpins are structurally conserved and thus the high level of similarity between the fourth repeat of VDAC and the C-terminal strands of BcsC is not the result of structural constraints.