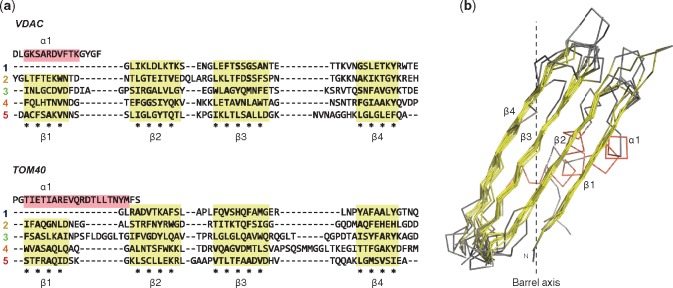
Fig. 4.
—Sequence and structural conservation of the repeat units from VDAC and TOM40. (a) Structure-based sequence alignment of the repeats identified in the HMM-consensus sequence of VDAC and TOM40 with HHrepID. Repeats were mapped onto their reference structure (TOM40: 5o8o_A; VDAC: 4c69_X), and (b) the structural superposition of the corresponding double ββ-hairpins was carried out with TMalign and manually adjusted using UCSF Chimera (Pettersen et al. 2004) without considering the N-terminal helix and the loop regions. In (a), the position and boundaries of the helices and strands, as of their reference structure, are shaded red and yellow, respectively; asterisks mark strand positions facing the outside of the barrel, with those in bold depicting hydrophobic, aromatic or small residues. Each sequence repeat is composed of two ββ-hairpins, with the exception of the first repeat, where the first strand appears to have been changed to an α-helix. All these double ββ-hairpins have a closely matching structure. In (b), a dashed line represents the transmembrane axis of the reference barrels, highlighting the strand tilt with respect to the membrane.