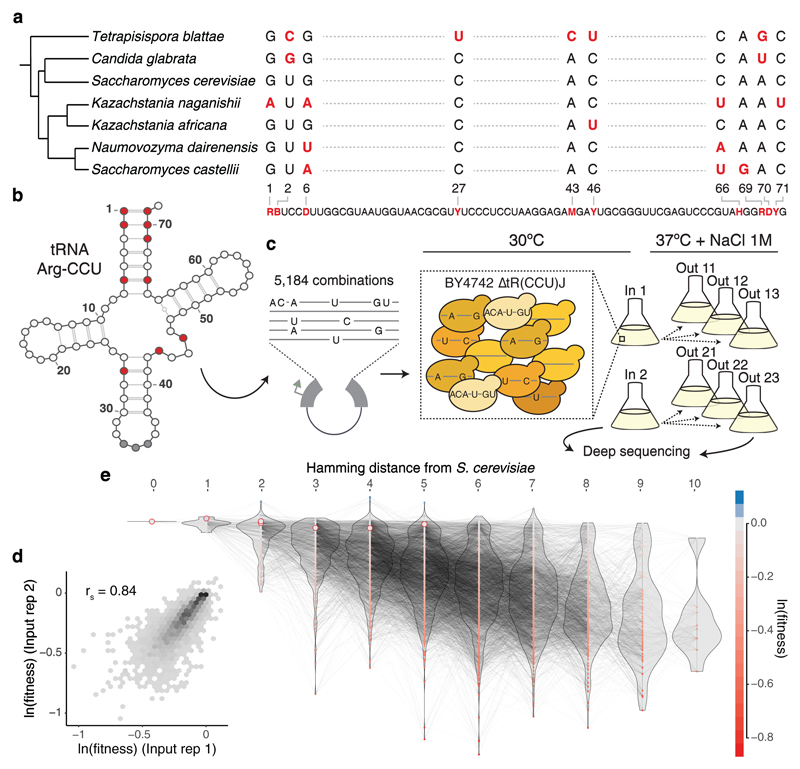
Figure 1. Combinatorially-complete fitness landscape of a tRNA.
a, Species phylogenetic tree26 and multiple sequence alignment of the tRNA-Arg-CCU orthologs. Shown variable positions across the seven yeast species with the synthesized library below. R - A or G, B - C, G or T, D - A, G or T, Y - C or T, M- A or C, H - A, C or T. b, Secondary structure of S. cerevisiae tRNA-Arg-CCU (varied positions in red). c, Selection experiment and structure of the replicates. From each independent yeast transformation (input) three independent selection experiments were performed. d, Correlation between weighted-averaged input replicates (rs = Spearman correlation coefficient, n = 4,176 genotypes). e, Fitness landscape of the tRNA-Arg-CCU genotipes (nodes). Colour indicates ln(fitness) relative to the S. cerevisiae tRNA. Edges connect genotypes differing by a single substitution. Genotypes and distribution of fitness values (violins) are arranged in the x-axis according to the total number of substitutions from the S. cerevisiae tRNA. Highlighted nodes indicate the genotypes of the seven extant species.