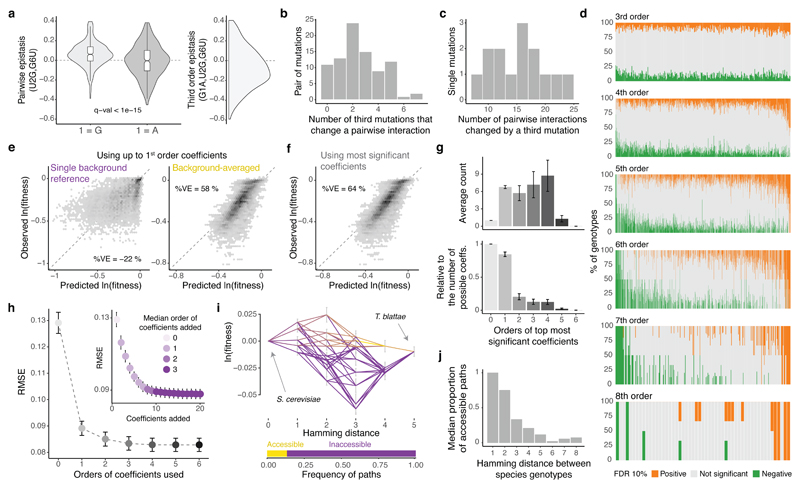
Figure 4. Averaging coefficients across genetic backgrounds and using higher order epistatic terms is important for genetic prediction.
a, Changes in the distribution of pairwise epistasis when the genetic backgrounds contain or not the indicated mutation (left) and distribution of the corresponding third order epistasis values (right). b, Distribution of pairwise interactions altered by a third mutation. c, Distribution of single mutations involved in a third order interaction. d, Proportion of genetic backgrounds in which each combination of mutations from 3rd to 8th order interact positively (orange) or negatively (green) at a FDR<0.1. e, Agreement between observed and predicted fitness values of all 8th order complete sub-landscapes (n = 19,456 genotypes, 76 sub-landscape with 256 genotypes each) when using up to 1st order epistatic coefficients, relative to a single background genotype (left) or averaged across backgrounds (right, 10-fold cross-validation). %VE = Percentage of variance explained. f, Agreement between observed and predicted fitness values for all complete 8th order sub-landscapes when using the most significant epistatic coefficients, estimated by 10-fold cross-validation. g, Mean root-mean-square error (RMSE) across the 76 8th order sub-landscapes when cumulatively adding most significant coefficients determined by cross-validation (right, colour indicates de median order of the coefficient added across 76 sub-landscapes) or all significant coefficients from the same order (left). Error bars are 95% CI. h, Mean orders of most significant epistatic coefficients (top, absolute counts; bottom, relative to the possible number of coefficients per order). Error bars are 95% CI. i, Example of shortest paths between two extant species (top) and accessible proportion (bottom). j, Average frequency of accessible paths between species.