Abstract
Elucidating gene expression programs within a cell-specific manner is a grand challenge for biologists. Harder still is the abilty to have kinetic control over such experiments. Metabolic labeling with bioorthogonally-functionalized metabolic intermediates provides a means to profile RNA expression in a cell-specific manner, but there is still lack of kinetic resolution. Herein we present the synthesis and evaluation of photocaged metabolic uracil intermediates. We compare the photo-decaging properties and demonstrate their utility in metabolic labeling experiments in a cell-specific manner. We anticipate our approach will have far-reaching impact as they provide control over tagging of nascent RNA.
Characterizing the biological profile of cells often relies on a complete description of the RNA content, or transcriptome.1, 2 The key challenge for characterizing nascent RNA from the steady-state cellular pool is to enrich newly transcribed RNA for downstream analyses such as qRT-PCR or sequencing. One method to enrich an RNA pool is to install RNA with bioorthogonal chemical handles that permit selective enrichment. As such, the time after the addition of the analog serves as a time-stamp of enriched RNA. Several accounts have shown that cells can be incubated with alkynyl and/or azido modified nucleosides, which are then converted to modified nucleoside triphosphates for eventual incorporation into RNA and DNA.3-8 These studies have demonstrated that introducing such metabolic intermediates is a robust method for labeling and enriching the nascent cellular RNA.
More recently, the bioorthogonal approaches have been expanded to become cell-specific. In these accounts, cells of interest express an enzyme that helps incorporate an inert bioorthogonal nucleoside intermediate into cellular RNA. For example, modified uracils can be converted to 5′-phosphoryluridines by exogenous expression of the enzyme uracil phosphoribosyltransferase (UPRT).9-11 “Uncaging” a modified nucleoside with a blocking group is an equally useful cell-type specific strategy for labeling nascent RNA pools. One example is the liberation of a protected 2′-azidoadenosine by the enzyme penicillin G amidase (PGA).12 The key observation from these studies is that modified nucleoside intermediates can be chemically controlled by functional group placement. These intermediates can remain inert until being transformed into a metabolic-active form by functional group removal.
The aforementioned studies have paved the way for robust chemical approaches to nascent metabolic labeling of cellular RNA. Nevertheless, there still is a critical issue of such experiments that needs to be addressed: lack of kinetic control. The ability to turn on metabolic labeling inside cells with defined temporal control is even more important in more complex environments like whole animals. This is because such experiments are performed by long incubation times of analogs to permit tissue perfusion to cells of interest. During this time analogs are being incorporated into RNA and as such the timing of RNA profiling is blurred and the resolution is lost. For example, modified nucleoside approaches have been utilized in C. Elegans, Zebrafish, and even mice. In most of these reports, long incubation times (>8 hours) of the nucleosides were used to infuse the animal for metabolic labeling.9, 10, 13, 14 An attractive alternative would be to permit analog diffusion without RNA incorporation. Kinetic control, permitted by controlled release of the RNA metabolic intermediate would afford kinetic control as to when the analog is incorporated into RNA – a major improvement in animal-based RNA tracking and labelling. This will continue to be a major weakness in the metabolic labeling approach unless more sophisticated experimental protocols are developed to control the time of initiation.
Herein we approach this challenge with the concept of using light to “uncage” a bioorthogonal nucleoside (Fig. 1A). We systematically test the design and “uncaging” of several uracil analogs, comparing the differences in photo-liberation of blocking groups. We also show that our approach works in cells to liberate uracil to be eventually incorporated into RNA. Lastly, we take advantage of our recently reported 5-ethynyluracil (5EU) / UPRT pair to convert an “uncaged” 5EU for kinetic control over cell-specific metabolic labeling of RNA (Fig. 1B). As the UPRT system has been widely used inside living animals10, we anticipate our results will be of wide interest to the growing field of nascent RNA expression analysis in vivo.
Fig. 1. Photo-controlled metabolic labeling of RNA.
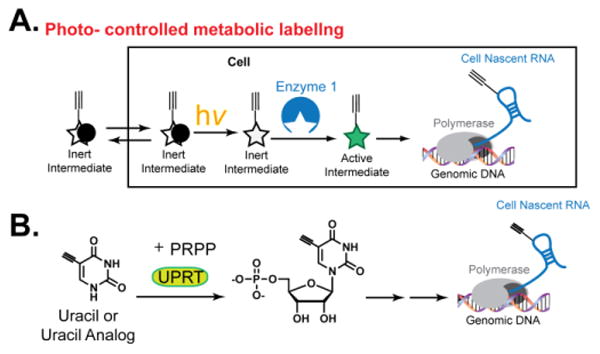
(A) Schematic of photo-controlled cell-specific metabolic labeling of RNA. (B) Schematic of UPRT-dependent RNA labeling.
Following our recent observation that a 5EU / UPRT pair can metabolically label RNA in a cell-specific manner, we began to reason how we could perturb the interaction between 5EU and UPRT. Inspection of the co-crystal structure of uracil-bound UPRT revealed that the N3-position of uracil is packed tightly up against amino acids in the binding site (Fig. 1C). Additionally, the N1 nitrogen is in a more solvent exposed point (Fig. 1C); however, this is the sight of glycosidic bond formation and as such would be an inhibitor of the eventual reaction.
We began by synthesizing a 2-nitrobenzyl caged uracil (Cmpd 1, Fig. 2A, ESI†). 2-nitrobenzyl caged compounds have been used for uncaging small molecules in cells and even in vivo.15, 16 We tested photolability of the nitrobenzyl group by exposure to 365nm light, observing the kinetics of uncaging by 1H NMR. Unfortunately, even after one hour of light exposure, there was undetectable uncaging (Fig. S1, ESI†). We reasoned that the high pKa of the N1 nitrogen (pKa = 10) renders this nitrogen a weak leaving group and as such the uncaging reaction would not proceed at reasonable rate and yield. We then synthesized a modified nitrophenyl version (6-nitropiperonyloxymethyl, NPOM), which contains a methylene carbon inserted between the aryl protecting group and N1, to improve the uncaging process (Cmpd 2, Fig. 2A, ESI†). NPOM deprotection has been used on pyridine nucleobases for efficient deprotection in RNA sequences and DNA plasmids in vivo.17, 18 Exposure to 365nm light and following the reaction by HPLC showed that the yield of uncaging approached 50% after 20 minutes of light exposure (Fig. 2B). Seeking to further improve the uncaging yield within a similar time window we synthesized the N3 derivative (Cmpd 3, Fig. 2A, ESI†); we reasoned that the slightly lower pKa (pKa = 9) and lower nucleophilicity of the N3 would render the uracil more labile to deprotection. Consistent with this notion, HPLC demonstrated that the yield of uncaging after 20 minutes of exposure increased by 20% (∼70%; Fig. S2, ESI†). These results demonstrate that the position of photocages on uracil, as well as the structure of protecting groups used, are critical parameters to optimize when designing protected nucleobases.
Fig. 2. Testing photo-“uncaging” of uracil analogs.
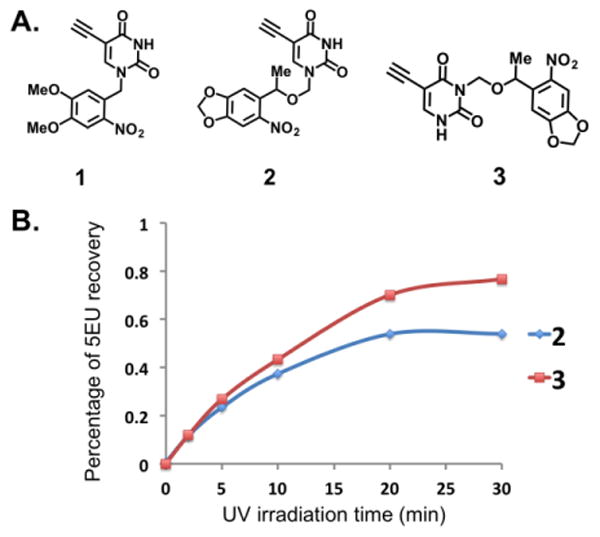
(A) Structures of synthesized analogs. (B) HPLC results demonstrating successful un-“caging” of analogs 2 and 3.
After identifying our best photocaged uracil, we switched our focus to cell studies to determine if the protected uracil analogs would be incorporated into RNA upon light exposure. We used a Northern blot analysis, which demonstrates RNA labelling through biotinylation, with an azido biotin, after a copper catalyzed cycloaddition (CuAAC) on isolated total RNA (Fig. 3A). As shown in Fig. 3B (Fig. S3, ESI†), incubation of cells with 3 resulted in undetectable RNA incorporation. In contrast, when cells were incubated with 3, and exposed to light for 20 minutes, robust RNA incorporation was observed after 5 hours by Northern blot (Fig. 3B, Fig. S3, ESI†). Cells not expressing UPRT, but exposed to light did not result in analog incorporation. These results demonstrate that caged uracil is incorporated into cellular RNA, only when UPRT-expressing cells are exposed to light.
Fig. 3. Testing photo-controlled incorporation of 5EU into cellular RNA.
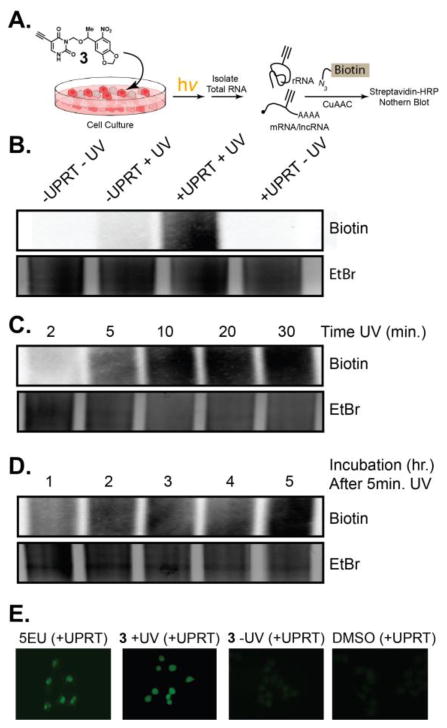
(A) Schematic of experiments used to test incorporation. (B) Northern blot demonstrating specificity of “uncaged” 5EU incorporation into RNA. (C) Northern blot testing UV light exposure times. (D) Northern blot testing time of incubation after 5 minutes of UV exposure. (E) Fluorescence imaging for incorporation of 5EU in living cells.
We further worked to test the limitations of the conditions for incubation and light exposure. First we tested how much time of light exposure was necessary to observe robust incorporation of liberated 5EU. Fig. 3C (Fig. S3, ESI†) shows that even just 5 minutes of light exposure, followed by incubation with cells for 5 hours, results in robust signal by Northern blot. Fig. 3D (Fig. S3, ESI†) demonstrates that 5-min light exposure and one hour incubation also results in significant signal for 5EU incorporation into RNA. The time of incubation of un-“caged” 3 are similar to the results we have observed with UPRT-dependent RNA labelling with 5EU.12 These results further demonstrate the ability of 5EU to be caged and efficiently liberated by light exposure. Further, they support the notion that once 5EU is liberated it is incorporated into cellular RNA with similar kinetics of just 5EU.
Cellular imaging is a widely used method for testing incorporation of exogenous analogs into cellular metabolic pathways. Uracil and other pyrimidine analogs have been shown to primarily locate within the nucleolus of cells. This is due to the abundant synthesis and cellular content of ribosomal RNA. After 5EU incubation, the majority of signal comes from the nucleolus, consistent with our previous reports.12 Incubation of UPRT expressing cells with 3 alone resulted in no cellular imaging signals (Fig. 3E). Incubation of UPRT expressing cells with 3, followed by light exposure resulted in robust imaging signals inside the nucleus, in similar fashion as 5EU. Overall, these results further demonstrate that photo-uncaging of modified uracil analogs can endow kinetic control over metabolic labelling of RNA, in a cell-specific manner.
Herein we have presented the first report of photo-controlled metabolic biomacromolecule labeling in a cell-specific manner. We have also done a systematic evaluation of protecting group design and placement for controlled release of uracil into medium. Metabolic labeling of RNA has found utility in describing nascent RNA synthesis, the rate of RNA decay, and even RNA localization. As such, the ability to have kinetic control over RNA metabolic labeling would improve the resolution of nascent RNA capture, decay rate analysis and even RNA location characterization. The widespread use of photo-controlled reaction in vitro and even in living animals further underscores the importance of our results. We anticipate many labs will adopt such a strategy to have better control over nascent RNA synthesis and downstream characterization.
Supplementary Material
Acknowledgments
We thank members of the Spitale lab for critical reading of the manuscript. RNA research in the Spitale lab is supported by startup funds through the University of California, Irvine and the NIH Director's New Innovator Award (1DP2GM119164).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Mol Cell. 2017;65:631–643 e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 2.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng Y, Beal PA. Bioorg Med Chem Lett. 2016;26:1799–1802. doi: 10.1016/j.bmcl.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jao CY, Salic A. Proc Natl Acad Sci U S A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curanovic D, Cohen M, Singh I, Slagle CE, Leslie CS, Jaffrey SR. Nat Chem Biol. 2013;9:671–673. doi: 10.1038/nchembio.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nainar S, Beasley S, Fazio M, Kubota M, Dai N, Correa IR, Jr, Spitale RC. Chembiochem. 2016;17:2149–2152. doi: 10.1002/cbic.201600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neef AB, Pernot L, Schreier VN, Scapozza L, Luedtke NW. Angew Chem Int Ed Engl. 2015;54:7911–7914. doi: 10.1002/anie.201500250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neef AB, Luedtke NW. Chembiochem. 2014;15:789–793. doi: 10.1002/cbic.201400037. [DOI] [PubMed] [Google Scholar]
- 9.Chatzi C, Zhang Y, Shen R, Westbrook GL, Goodman RH. eNeuro. 2016;3 doi: 10.1523/ENEURO.0024-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Cleary MD, Stankunas K, Doe CQ. Genes Dev. 2013;27:98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MR, Robinson KJ, Cleary MD, Doe CQ. Nat Methods. 2009;6:439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen K, Fazio M, Kubota M, Nainar S, Feng C, Li X, Atwood SX, Bredy TW, Spitale RC. J Am Chem Soc. 2017;139:2148–2151. doi: 10.1021/jacs.6b11401. [DOI] [PubMed] [Google Scholar]
- 13.Jungkamp AC, Stoeckius M, Mecenas D, Grun D, Mastrobuoni G, Kempa S, Rajewsky N. Mol Cell. 2011;44:828–840. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson T, Nicolson T. BMC Genomics. 2015;16:842. doi: 10.1186/s12864-015-2072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Koo SC, Li NS, Piccirilli JA. Nucleosides Nucleotides Nucleic Acids. 2015;34:114–129. doi: 10.1080/15257770.2014.965256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaplovsky M, Il'ichev YV, Kamdzhilov Y, Kombarova SV, Mac M, Schworer MA, Wirz J. Photochem Photobiol Sci. 2005;4:33–42. doi: 10.1039/b409927c. [DOI] [PubMed] [Google Scholar]
- 17.Govan JM, Young DD, Lusic H, Liu Q, Lively MO, Deiters A. Nucleic Acids Res. 2013;41:10518–10528. doi: 10.1093/nar/gkt806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Deiters A. Acc Chem Res. 2014;47:45–55. doi: 10.1021/ar400036a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


