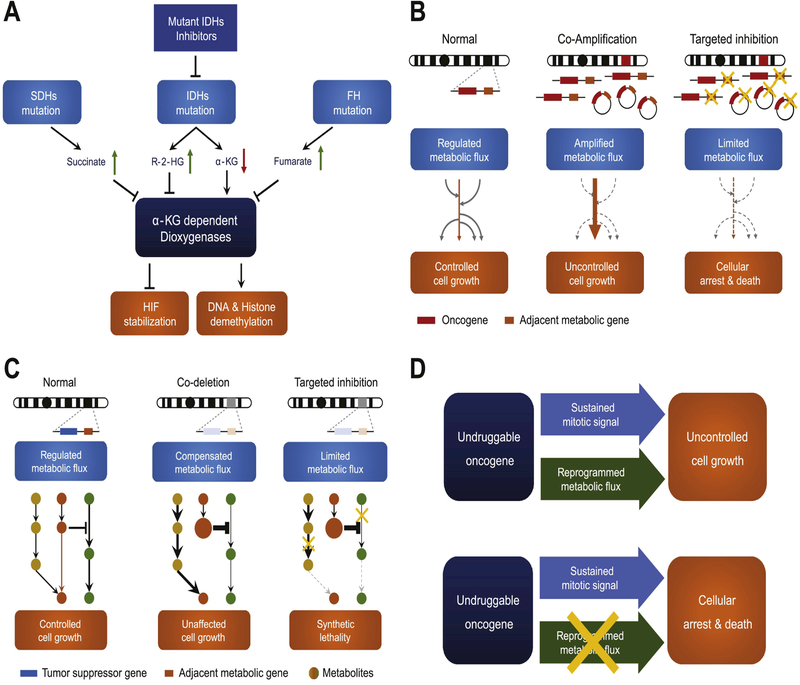
Fig. 3. Targeting metabolic co-dependency in cancer.
(A) Mutations of SDHs and FH genes lead to accumulation of their individual substrates succinate and fumarate. Mutated IDHs acquire new function to convert α-KG into R-2-HG. These metabolites can inhibit α-KG dependent dioxygenases, and subsequently abolish HIF de-stabilization and DNA, histone demethylation. (B) When a metabolic gene recurrently co-amplified with a nearby oncogene, either in the form of chromosomal or extrachromosomal DNA, can drive the metabolic flux to a specific direction to aid malignant transformation, creating a metabolic co-dependency. Hitting co-amplified metabolic gene is a potential strategy to halt cancer cells. (C) Metabolic flux in normal cells comprises parallel or redundant pathways with feedback regulation mechanism, allowing metabolic plasticity. Metabolic gene co-deletion with a tumor suppressor impairs the plasticity, creating the potential to exploit synthetic lethality. (D) Oncogenes confer uncontrolled cell growth by providing sustained mitotic signal and reprogramming metabolism. Targeting reprogrammed metabolism becomes an alternative strategy when oncogene is not ideally druggable.